
ʻO ka Genome Assembly ma muli o Hi-C
Pono lawelawe
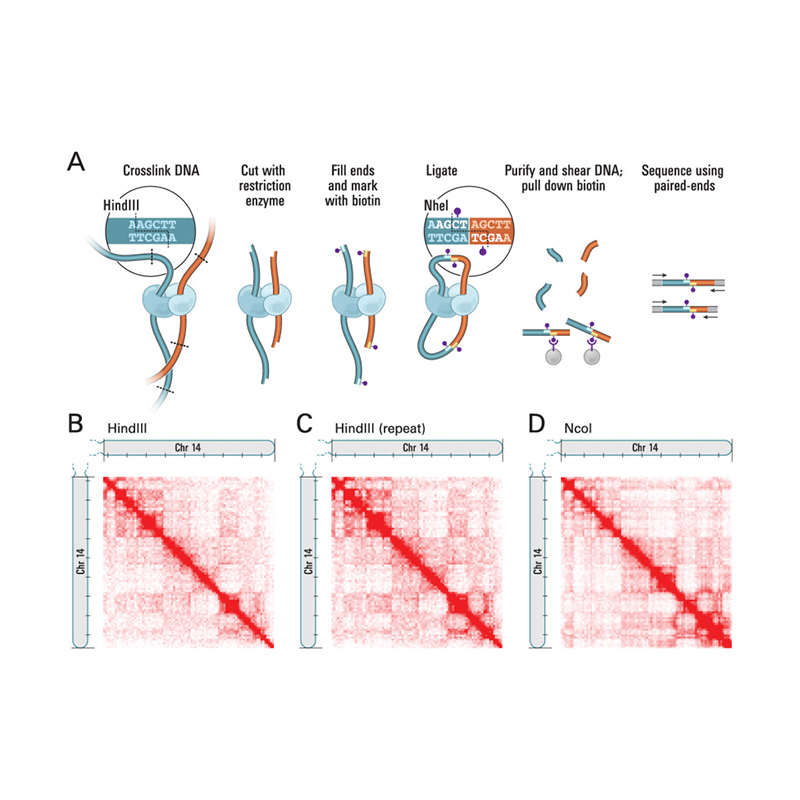
Nānā nui o Hi-C
(Lieberman-Aiden E et al.,ʻepekema, 2009)
● ʻAʻole pono ke kūkulu ʻana i ka heluna kanaka no ka hoʻopaʻa ʻana;
● Higher marker density e alakaʻi ana i ke kiʻekiʻe contigs anchoring ratio ma luna o 90%;
● Hiki i ka loiloi a me ka hoʻoponopono ʻana i nā hui genome e kū nei;
● ʻOi aku ka pōkole o ka manawa huli me ka ʻoi aku ka pololei o ka hui genome;
● Nui ka ʻike me 1000 mau hale waihona puke Hi-C i kūkulu ʻia no 500 mau ʻano;
● ʻOi aku ma mua o 100 mau hihia kūleʻa me ka helu ʻana i hoʻopuka ʻia ma luna o 760;
● ʻO ka hui genome ma muli o Hi-C no ka genome polyploid, 100% ka helu heleuma i loaʻa i ka papahana mua;
● ʻO nā patent a me nā lako polokalamu lako polokalamu no nā hoʻokolohua Hi-C a me ka ʻikepili ʻikepili;
● Ua hoʻomohala ʻia ka polokalamu hoʻoponopono ʻikepili i hoʻomohala ʻia, hiki ke hoʻoneʻe i ka poloka manual, hoʻohuli, hoʻopau a hana hou.
Nā kikoʻī lawelawe
|
ʻAno waihona
|
Papahana | Heluhelu Length | Manaʻo Kūkākūkā |
| Hi-C | ʻO Illumina NovaSeq | PE150 | ≥ 100X |
Nā loiloi bioinformatics
● Ka mana o ka maikaʻi o ka ʻikepili maka
● Ka mana o ka hale waihona puke Hi-C
● hui genome ma muli o Hi-C
● loiloi ma hope o ka hui ʻana
Nā Koina Laʻana a me ka Hoʻouna ʻana
Nā Koina Laʻana:
| Holoholona | ʻāhuʻu | Nā mea kanu
|
| ʻO ka ʻiʻo maloʻo: 1-2g no ka waihona Nā pūnaewele: 1x 10^7 pūnaewele no ka waihona | ʻiʻo paʻa hau: 1g no ka waihona | ʻO ka ʻiʻo maloʻo: 1-2g no ka waihona
|
| * Manaʻo ikaika mākou e hoʻouna ma kahi liʻiliʻi 2 aliquots (1 g kēlā me kēia) no ka hoʻokolohua Hi-C. | ||
Manaʻo ʻia ka hāʻawi laʻana
Pahu: 2 ml centrifuge tube (ʻAʻole ʻōlelo ʻia ʻo Tin foil)
No ka hapa nui o nā laʻana, paipai mākou ʻaʻole e mālama i loko o ka ethanol.
Laʻana hōʻailona: Pono e hōʻailona ʻia nā laʻana a e like me ka palapala ʻike hāpana i waiho ʻia.
Hoʻouna: Ka hau maloʻo: Pono e hoʻopili mua ʻia nā laʻana i loko o nā ʻeke a kanu ʻia i loko o ka hau maloʻo.
Holo Hana Hana

Hoʻolālā hoʻokolohua

Hāʻawi laʻana

Ka unuhi ʻana o DNA

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili

Nā lawelawe ma hope o ke kūʻai aku
*ʻO nā hopena demo i hōʻike ʻia ma ʻaneʻi mai nā genome i paʻi ʻia me Biomarker Technologies
1.Hi-C ka launa pū wela palapala 'āina oCamptotheca acuminatagenome.E like me ka mea i hōʻike ʻia ma ka palapala ʻāina, pili ʻole ka ikaika o nā pilina me ka mamao laina, e hōʻike ana i kahi hui pae chromosome pololei loa.(Ka lākiō hele: 96.03%)
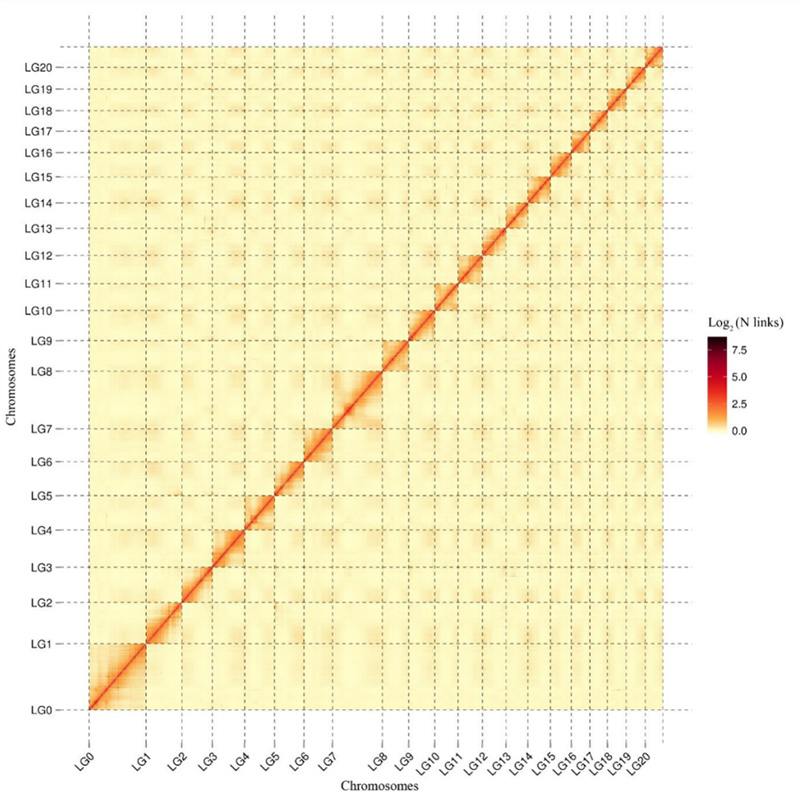
Kang M et al.,Nature Communications, 2021
2.Hi-C maʻalahi i ka hōʻoia o inversions ma waenaʻO Gossypium hirsutumL. TM-1 A06 aG. arboreumChr06
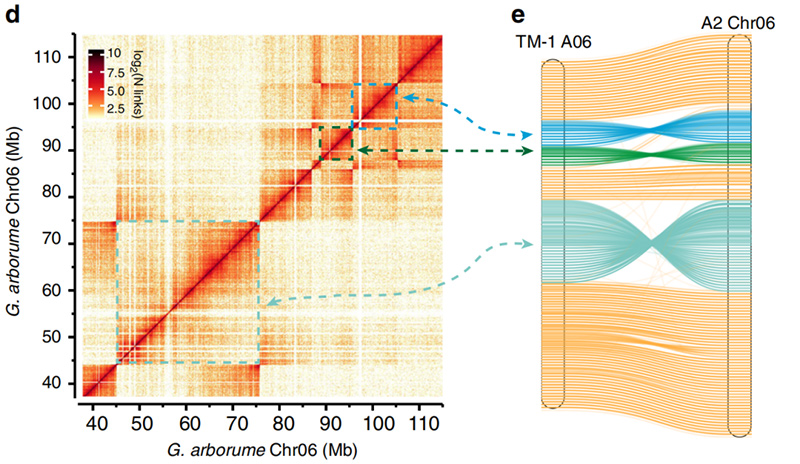
ʻO Yang Z et al.,Nature Communications, 2019
3.Assembly a biallelic differentiation o ka cassava genome SC205.Hōʻike ʻia ka māhele wela Hi-C i nā chromosomes homologous.
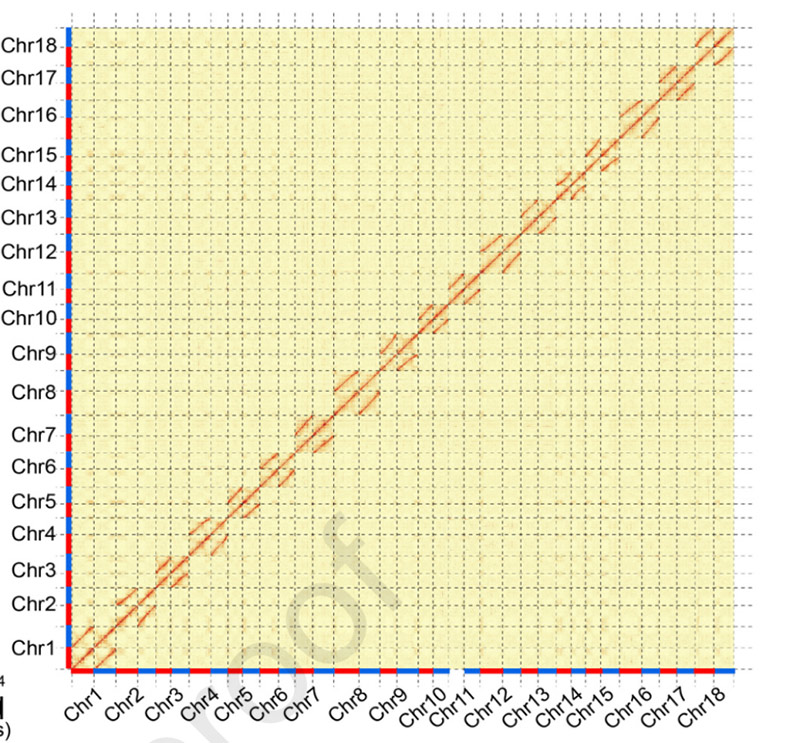
Hu W et al.,Mea Molekula, 2021
4.Hi-C heatmap ma ʻelua hui genome ʻano ʻano Ficus:F.microcarpa(ka lākiō heleuma: 99.3%) aF.hispida (ka lākiō heleuma: 99.7%)
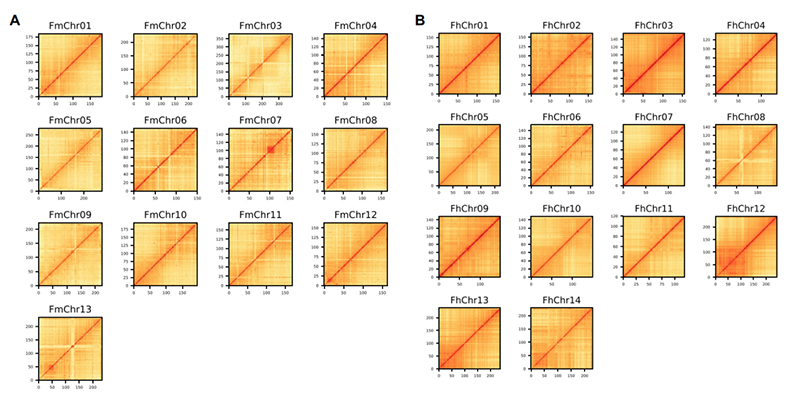
Zhang X et al.,Kelepona, 2020
Hihia BMK
Hāʻawi ʻo Genomes o ka lāʻau Banyan a me Pollinator Wasp i nā ʻike i ka Fig-wasp Coevolution
Paʻi ʻia: Kelepona, 2020
Hoʻolālā kaʻina hana:
F. microcarpa genome: Ma kahi.84 X PacBio RSII (36.87 Gb) + Hi-C (44 Gb)
F. hispidagenome: Ma kahi.97 X PacBio RSII (36.12 Gb) + Hi-C (60 Gb)
Eupristina verticillatagenome: Ma kahi.170 X PacBio RSII (65 Gb)
Nā hopena koʻikoʻi
1. Ua kūkulu ʻia ʻelua genomes lāʻau banyan a me hoʻokahi genome pollinator wasp me ka hoʻohana ʻana iā PacBio sequencing, Hi-C a me ka palapala pili.
(1)F. microcarpagenome: Ua hoʻokumu ʻia kahi hui o 426 Mb (97.7% o ka nui genome i manaʻo ʻia) me ka contig N50 o 908 Kb, BUSCO helu o 95.6%.Ma ka huina o 423 Mb kaʻina i hoʻopaʻa ʻia i 13 chromosomes e Hi-C.He 29,416 nā genes-coding protein.
(2)F. Hispidagenome: ʻO kahi hui o 360 Mb (97.3% o ka nui o ka genome i manaʻo ʻia) ua loaʻa me ka contig N50 o 492 Kb a me ka helu BUSCO o 97.4%.He 359 Mb mau kaʻina i hoʻopaʻa ʻia ma nā chromosomes 14 e Hi-C a ua like loa me ka palapala ʻāina pili kiʻekiʻe.
(3)Eupristina verticillatagenome: Ua hoʻokumu ʻia kahi hui o 387 Mb (Ka nui genome i manaʻo ʻia: 382 Mb) me ka contig N50 o 3.1 Mb a me ka helu BUSCO o 97.7%.
2. Hōʻike ka hoʻohālikelike ʻana i nā genomics i ka nui o nā ʻano like ʻole ma waena o ʻeluaFicusnā genomes, ka mea i hāʻawi i nā kumuwaiwai waiwai nui no nā haʻawina evolution adaptive.ʻO kēia haʻawina, no ka manawa mua, hāʻawi i nā ʻike i ka coevolution Fig-wasp ma ka pae genomic.
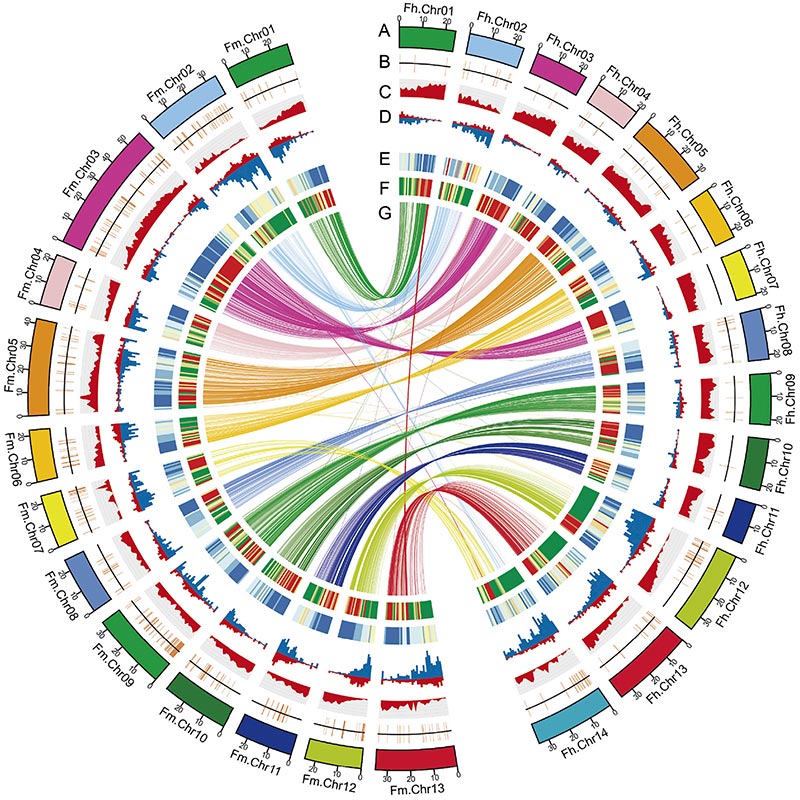 Kiʻi Circos ma nā hiʻohiʻona genomic o ʻeluaFicusnā genomes, me nā chromosomes, nā duplications segmental (SDs), nā transposon (LTR, TEs, DNA TEs), ka hōʻike gene a me ka synteny | 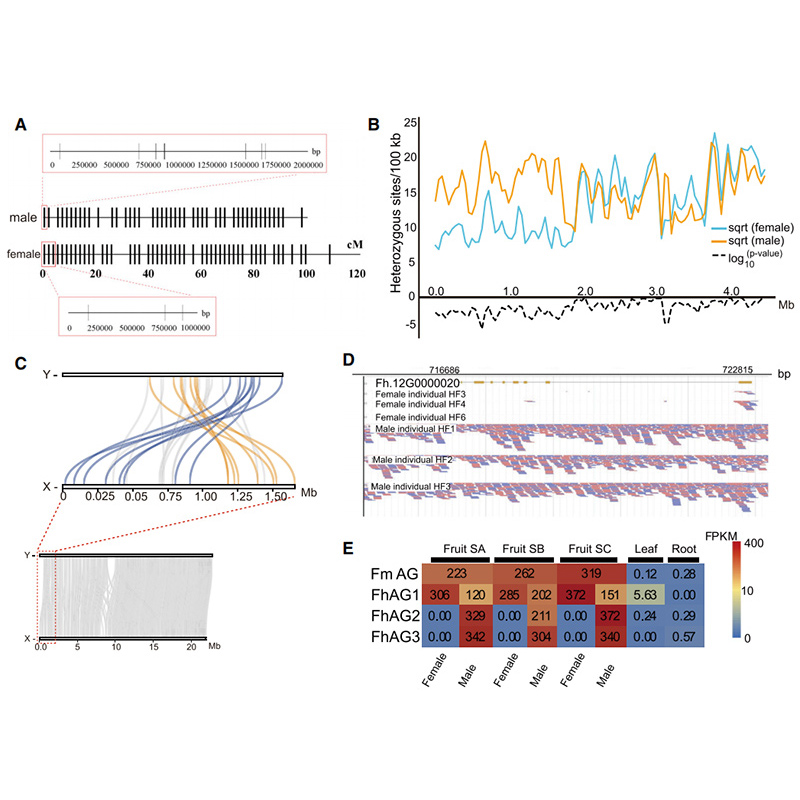 Ka ʻike ʻana o ka Y chromosome a me ka gene moho hoʻoholo kāne |
Zhang, X. , et al."Hāʻawi nā Genomes o ka lāʻau Banyan a me Pollinator Wasp i nā ʻike i ka Fig-Wasp Coevolution."Pūnaewele 183.4(2020).














