GENOME EVOLUTION

Comparative genome analyses highlight transposon-mediated genome expansion and the evolutionary architecture of 3D genomic folding in cotton
Nanopore sequencing | Hi-C | PacBio sequencing | Illumina | RNA-sequencing | 3 D genome architecture | Transposon | Comparative genomics
In this study, Biomarker Technologies provided technical support on Nanopore sequencing, Hi-C and relevant bioinformatic analysis.
Abstract
Transposable element (TE) amplification has been recognized as a driving force mediating genome size expansion and evolution, but the consequences for shaping 3D genomic architecture remains largely unknown in plants. Here, we report reference-grade genome assemblies for three species of cotton ranging three-fold in genome size, namely Gossypium rotundifolium (K2), G. arboreum (A2), and G. raimondii (D5), using Oxford Nanopore Technologies. Comparative genome analyses document the details of lineage-specific TE amplification contributing to the large genome size differences (K2, 2.44 Gb; A2, 1.62 Gb; D5, 750.19 Mb), and indicate relatively conserved gene content and synteny relationships among genomes. We found that approximately 17% of syntenic genes exhibit chromatin status change between active (“A”) and inactive (“B”) compartments, and TE amplification was associated with the increase of the proportion of A compartment in gene regions (~ 7,000 genes) in K2 and A2 relative to D5. Only 42% of topologically associating domain (TAD) boundaries were conserved among the three genomes. Our data implicate recent amplification of TEs following formation of lineage-specific TAD boundaries. This study sheds light on the role of transposon-mediated genome expansion in the evolution of higher-order chromatin structure in plants.
Key statistics of genome assembly
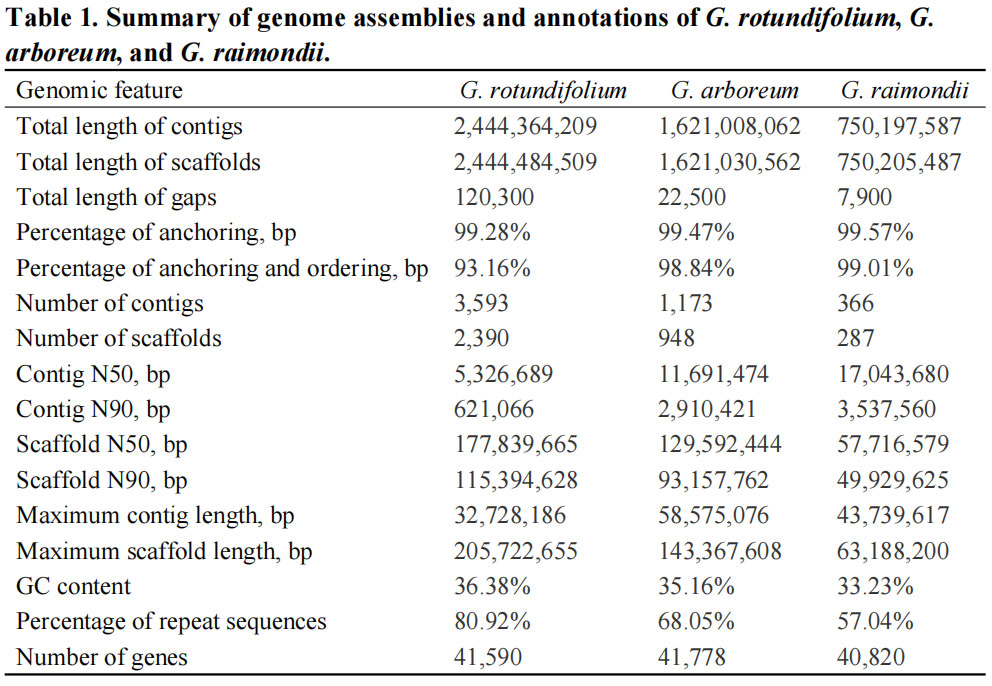
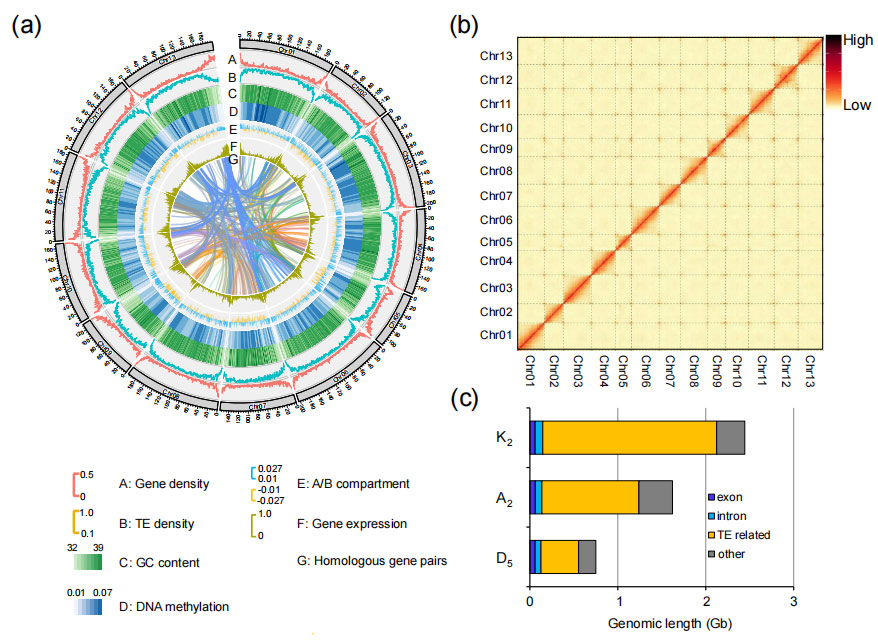
Figure. Genome assembly and feature description of G. rotundifolium (K2)
News and Highlights aims at sharing the latest successful cases with Biomarker Technologies, capturing novel scientific achievements as well as prominent techniques applied during the study.
Post time: Jan-05-2022

