GENOME EVOLUTION, PANGENOME
What is Pan-genome?
Accumulated evidences show that variance between different strains of a species can be huge. One single genome is far from enough to gain the whole picture of genetic information of a single species. The purpose of Pan-genome study is to obtain a more comprehensive genomic graph of a species and decode the relationships between traits and genetic codes by conducting genome de novo assembly of numerous strains, which allows a deeper and broader mining of the variations.
Trends of Pan-genome Study
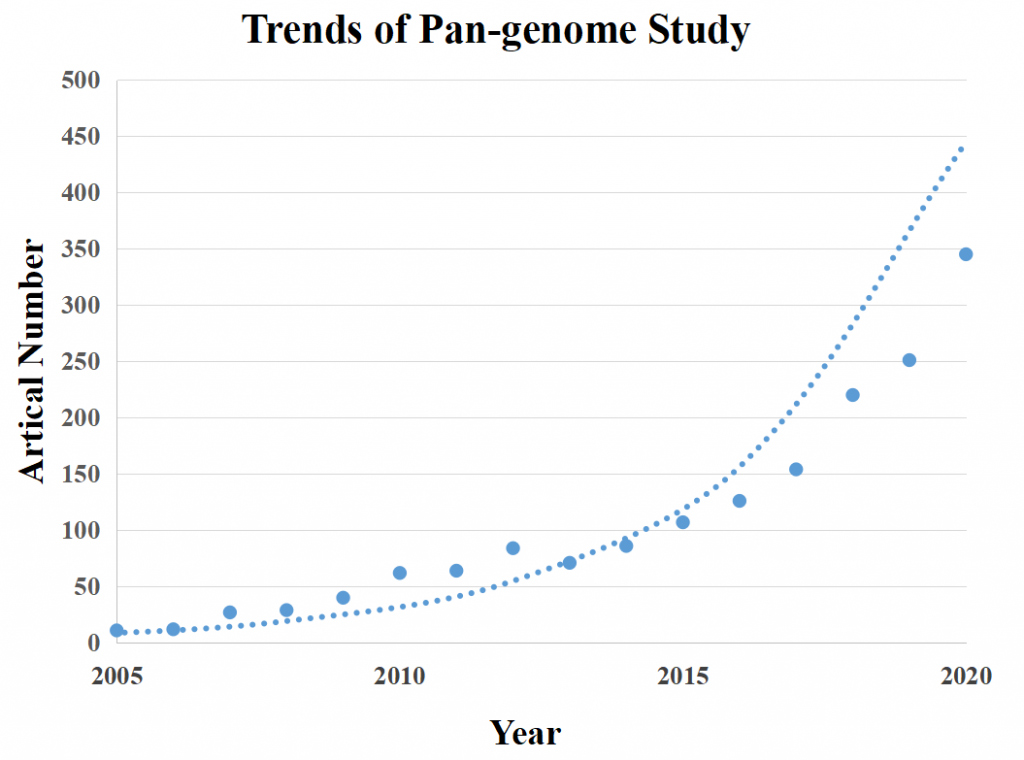
Figure 1 The trends of published research papers of Pan-genome.
Note: The figure shows the result of taking “pan-genome” as the key word to search the titles of articles published in Nature, Cell and Science series journals
a. Reads from multiple samples are aligned to a reference and unaligned ones are assembled into novel contigs. By adding these novel contigs to the original reference sequence, a pangenome reference can be constructed. Dispensable regions are determined based on mapping all reads back to the pangenome.
b. De novo assembly of the genomes of multiple accessions allows whole genome alignment approaches to identify dispensable genomic regions.
c. A pan-genome graph can be constructed from whole genome alignments or by de novo graph assembly, which efficiently stores variant information of dispensable regions as unique paths through the graph.
How to construct a Pan-genome?
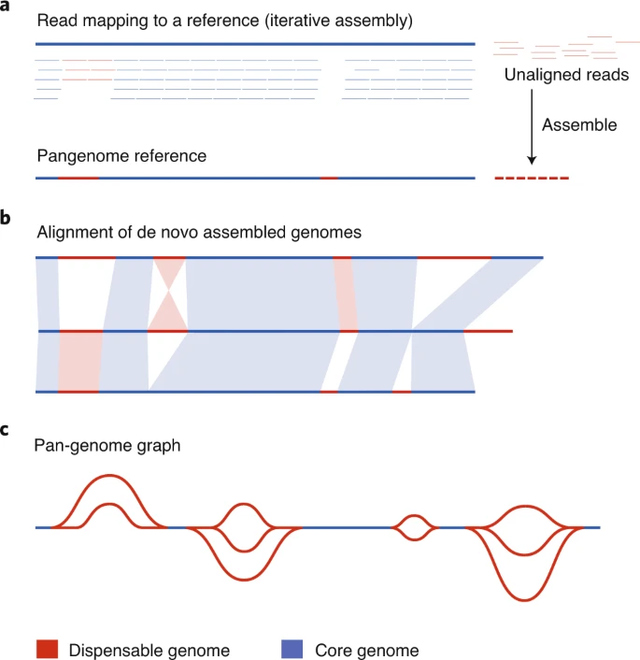
Figure 2 Comparison of pan-genome approaches 1
a. Reads from multiple samples are aligned to a reference and unaligned ones are assembled into novel contigs. By adding these novel contigs to the original reference sequence, a pangenome reference can be constructed. Dispensable regions are determined based on mapping all reads back to the pangenome.
b. De novo assembly of the genomes of multiple accessions allows whole genome alignment approaches to identify dispensable genomic regions.
c. A pan-genome graph can be constructed from whole genome alignments or by de novo graph assembly, which efficiently stores variant information of dispensable regions as unique paths through the graph.
Recently Published Pan-genomes
● Rape pan-genomes 2

● Tomato pan-genomes 3

● Rice Pan -genome 4

● Sunflower Pan-genome 5

● Soybean Pan-genome 6

● Rice Pan-genome 7

● Barley Pan-genome 8

● Wheat Pan-genome 9

● Sorghum Pan-genome 10

● Phytoplankton Pan-genome 11

Reference
1. Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D. Plant pan-genomes are the new reference. Nat Plants. 2020;6(8):914-920. doi:10.1038/s41477-020-0733-0
2. Song JM, Guan Z, Hu J, et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants. 2020;6(1):34-45. doi:10.1038/s41477-019-0577-7
3. Gao L, Gonda I, Sun H, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet. 2019;51(6):1044-1051. doi:10.1038/s41588-019-0410-2
4. Zhao Q, Feng Q, Lu H, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice [published correction appears in Nat Genet. 2018 Aug;50(8):1196]. Nat Genet. 2018;50(2):278-284. doi:10.1038/s41588-018-0041-z
5. Hübner S, Bercovich N, Todesco M, et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants. 2019;5(1):54-62. doi:10.1038/s41477-018-0329-0
6. Liu Y, Du H, Li P, et al. Pan-Genome of Wild and Cultivated Soybeans. Cell. 2020;182(1):162-176.e13. doi:10.1016/j.cell.2020.05.023
7. Qin P, Lu H, Du H, et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations [published online ahead of print, 2021 May 25]. Cell. 2021;S0092-8674(21)00581-X. doi:10.1016/j.cell.2021.04.046
8. Jayakodi M, Padmarasu S, Haberer G, et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature. 2020;588(7837):284-289. doi:10.1038/s41586-020-2947-8
9. Walkowiak S, Gao L, Monat C, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588(7837):277-283. doi:10.1038/s41586-020-2961-x
10. Tao Y, Luo H, Xu J, et al. Extensive variation within the pan-genome of cultivated and wild sorghum [published online ahead of print, 2021 May 20]. Nat Plants. 2021;10.1038 / s41477-021-00925-x. doi:10.1038/s41477-021-00925-x
11. Fan X, Qiu H, Han W, et al. Phytoplankton pangenome reveals extensive prokaryotic horizontal gene transfer of diverse functions. Sci Adv. 2020;6(18):eaba0111. Published 2020 Apr 29. doi:10.1126/sciadv.aba0111
Post time: Jan-04-2022

