KANAKA KANAKA
ʻano genetics
Hoʻomaopopo ka ʻōlelo lōʻihi i ka hoʻonui hou ʻana o GGC i NOTCH2NLC e pili ana me ka maʻi hoʻokomo intranuclear neuronal.
ONT resequencing |Ilumina |ʻO ke kaʻina exome holoʻokoʻa |ʻO CRISPR-Cas9 ONT i hoʻopaʻa ʻia i ke kaʻina hana |RNA-seq |ONT 5mC methylation kelepona
Nā mea nui
1.Ma ka loiloi Linkage ma kahi ʻohana NIID nui, ua ʻike ʻia ʻelua mau ʻāpana pili.
2.ONT-based long-read sequencing and Cas-9 mediated enrichment ONT sequencing found a potential genetic cause of NIID, GGC repeat expansions in 5′ UTR of NOTCH2NLC.Ua hōʻike ʻia kēia haʻawina i ka hoʻonui hou ʻana i nā genes kikoʻī kanaka no ka manawa mua i ulu aʻe ma o nā duplications segmental.
3. Ua hōʻike ʻia ke kaʻina ʻana o RNA i nā transcript antisense ʻino i ka hoʻomaka a i ʻole i loko o ka GGC e hoʻonui hou i nā wahi hoʻonui ma NOTCH2NLC.
Kāpae
NʻO ka euronal intranuclear inclusion disease (NIID) kahi maʻi neurodegenerative holomua a make, i hōʻike ʻia e ka hiki ʻana o ka eosinophilic hyaline intranuclear inclusions i loko o nā ʻōnaehana nerve waena a me peripheral.ʻO kāna mau hōʻike lapaʻau ʻokoʻa e hoʻonui i nā pilikia nui i ka ʻike a hiki i ka hoʻomaka ʻana o ka biopsy ʻili.Eia nō naʻe, ke hoʻomau nei nā ʻano hana histopathology i ka misdiagnosis, e koi ana i kahi ʻike genetic o NIID.
Nā mea i loaʻa
ʻIkepili Hoʻohui
SʻO ka hort-read sequencing e pili ana i ka genome sequencing holoʻokoʻa (WGS) a me ke kaʻina exome holoʻokoʻa (WES) i hana ʻia ma kahi ʻohana NIID nui (13 i hoʻopilikia ʻia a me 7 mau lālā i hoʻopilikia ʻole ʻia).Ua hōʻike ʻia ka loiloi loulou ma nā SNP i unuhi ʻia mai kēia mau ʻikepili i ʻelua mau wahi pili: kahi ʻāpana 3.5 Mb ma 1p36.31-p36.22 (LOD kiʻekiʻe = 2.32) a me kahi ʻāpana 58.1 Mb ma 1p22.1-q21.3 (LOD kiʻekiʻe: 4.21 ).Eia naʻe, ʻaʻole i ʻike ʻia nā SNP pathogenic a i ʻole CNV ma kēia mau wahi pili.
Hoʻonui hou GGC i NOTCH2NLC
NUa hoʻokō ʻia ke kaʻina hana anopore ma 13 i hoʻopilikia ʻia a me 4 mau lālā i hoʻopilikia ʻole ʻia mai nā ʻohana 8 (ʻo kekahi lālā i hoʻopilikia ʻia ua hoʻonohonoho ʻia e Pacbio lōʻihi heluhelu sequencing platform.).Hōʻike ka ʻikepili lōʻihi i ka maʻi e pili ana i ka hoʻonui hou ʻana o GGC i ka 5′ UTR o NOTCH2NLC gene mapping i 58.1 Mb i hoʻopili ʻia (Figure 1).Ua ʻike ʻia kēia mau hoʻonui hou ʻana i nā hihia NIID sporadic 40 a pau i hoʻāʻo ʻia e RP-PCR.
CUa hoʻohana ʻia ʻo as-9 i hoʻopaʻa ʻia i ka hoʻonohonoho ʻana ma ke kahua nanopore e hoʻokō i ka uhi heluhelu kiʻekiʻe ma ka NOTCH2NLC hana hou (100 X-1,795 X).Ua ʻae maikaʻi kēia mau ʻōlelo aʻoaʻo me nā ʻike mua ma ka hoʻonui hou ʻana o GGC.Eia hou, ua ʻike ʻia ka {(GGA)n (GGC)n}n i mea hōʻailona genetic no ka nāwaliwali-dominant phenotype (Figure 2).
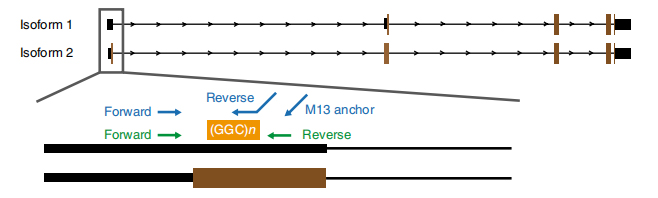
Kiʻi 1. Hoʻonui hou ʻia ka maʻi i ʻike ʻia ma ka exon 1 o NOTCH2NLC isoforms.
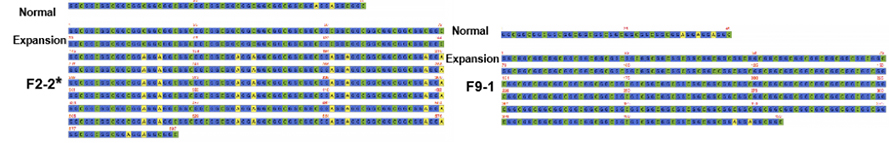
Kiʻi 2. ʻO nā kaʻina kuʻikahi o NPTCH2NLC e hana hou i nā maʻi NIID me (*) a i ʻole ka nāwaliwali-dominant phenotype
NʻO nā genes OTCH2NL nā genes kikoʻī kanaka, i manaʻo ʻia he kuleana koʻikoʻi i ka ulu ʻana o ka lolo kanaka a me nā maʻi neurological.Eia naʻe, ʻekolu mau genes pili ʻo NOTCH2 (NOTCH2NLA, NOTCH2NLB a me NOTCH2NLC) me> 99.1% ʻike ʻike ʻaʻole i hoʻoholo ʻia a hiki i ka hui genome kanaka hou loa.Ua hōʻike ʻo Synthesis-free a lōʻihi ka heluhelu ʻana ma ke kahua nanopore i nā mea maikaʻi loa i ka hoʻoponopono ʻana i nā wahi like kiʻekiʻe a (GGC) n hana hou me 100% GC-waiwai.
Hoʻonui hou GGC i NOTCH2NLC
TUa hoʻoponopono ʻia ke kaʻina hana ranscriptome ma 2 i hoʻopilikia ʻia a me 2 mau lālā i hoʻopilikia ʻole ʻia.Ua helu ʻia ka hohonu heluhelu maʻamau ma luna o nā kaula manaʻo a me nā antisense ma ke kahawai o nā exons mua o NOTCH2NL paralogs.Loaʻa nā transcripts anti-sense maʻamau i nā hihia i hoʻopilikia ʻia, e noho ana ma ka hoʻomaka a i ʻole i loko o ka ʻāina hoʻonui hou (Purple peaks ma F1-14 a me F1-16 ma Figure 3.).Eia hou, uaʻikeʻia nā 54 DEGs a ua hoʻonuiʻia nā mea a pau i nā hua'ōlelo GO a me MPO e pili ana i nā hana neuronal.
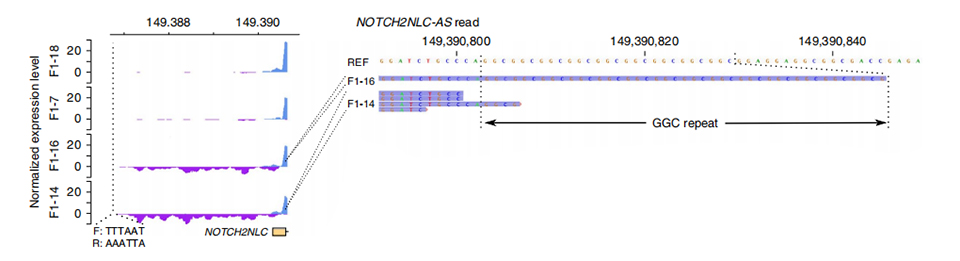
Kiʻi 3. Ka hohonu heluhelu maʻamau ma luna o ka exon mua o NOTCH2NLC i nā hihia i pili ʻole (ma luna) a i hoʻopili ʻia (ma lalo).
ʻenehana
ʻO Oxford Nanopore Teghnologies (ONT)
NHoʻokaʻawale ʻo anopore sequencing iā ia iho mai nā paepae hoʻonohonoho ʻē aʻe, no ka mea, heluhelu pololei ʻia nā nucleotides me ka ʻole o ke kaʻina hana synthesis DNA.I ka hele ʻana o kahi kaula DNA i loko o kahi pore protein nui nano (nanopore), hoʻopuka nā nucleotides ʻokoʻa i nā au ionic like ʻole, hiki ke hopu ʻia a hoʻololi ʻia i ke kaʻina o nā kumu.ʻAʻole hōʻike ʻo ONT sequencing platform i ka palena ʻenehana i ka lōʻihi o ka heluhelu DNA.No laila, loaʻa nā heluhelu Ultra-long (ULR) no ka hui genome o ke ʻano kiʻekiʻe.Eia kekahi, ʻo kēia mau heluhelu lōʻihi loa, ʻo ia ka lōʻihi e hiki ai ke hele i nā hiʻohiʻona kaʻina paʻakikī a i ʻole ka hoʻololi ʻana i ke ʻano, kōkua i ka pale ʻana i nā palena o ke kaʻina heluhelu pōkole ma aneʻi.

Nanopore sequencing
Hōʻike ʻano hoʻololi (SV).
SʻO ka ynthesis-free sequencing ka nui o ka mālama ʻana i ka ʻike DNA methylation ma ka template.Hoʻopuka ka Methylated A, T, C a me G i nā au ionic ʻokoʻa mai nā mea i hoʻokaʻawale ʻole ʻia, hiki ke heluhelu pololei ʻia e ka paepae.Hāʻawi ka Nanopore sequencing i ka hoʻopili ʻana i ka genome holoʻokoʻa o 5mC a me 6mA ma ka hoʻonā hoʻokahi-nucleotide.
Kuhikuhi
ʻO Jun Sone, et.al.Hōʻike ka ʻōlelo lōʻihi i ka hoʻonui hou ʻana o GGC i NOTCH2NLC e pili ana me ka maʻi intranuclear inclusion neuronal.Nature Genetics (2019)
ʻenehana a me nā mea koʻikoʻi manaʻo e kaʻana like i nā noi holomua hou loa o nā ʻenehana hoʻokaʻina kiʻekiʻe-throughput like ʻole ma nā kahua ʻimi noiʻi like ʻole a me nā manaʻo maikaʻi loa i ka hoʻolālā hoʻokolohua a me ka ʻimi ʻikepili.
Ka manawa hoʻouna: Jan-06-2022

