HUMAN GENOMICS
nature genetics
Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease
ONT resequencing | Illumina | Whole exome sequencing | CRISPR-Cas9 ONT targeted sequencing | RNA-seq | ONT 5mC methylation calling
Highlights
1.By Linkage analysis on a large NIID family, two linked regions were identified.
2.ONT-based long-read sequencing and Cas-9 mediated enrichment ONT sequencing discovered a potential genetic cause of NIID, GGC repeat expansions in 5′ UTR of NOTCH2NLC. This study reported repeat expansions in human-specific genes for the first time that evolved through segmental duplications.
3.RNA sequencing revealed abnormal antisense transcripts in the beginning or inside GGC repeat expansion regions in NOTCH2NLC.
Background
Neuronal intranuclear inclusion disease (NIID) is a progressive and fatal neurodegenerative disease, which is characterized by the presence of eosinophilic hyaline intranuclear inclusions in central and peripheral nervous systems. Its highly variable clinical manifestations raise great difficulties in diagnosis until introduction of skin biopsy. However, histopathology-based methods is still suffering from misdiagnosis, which is calling for a genetic understanding of NIID.
Achievements
Linkage Analysis
Short-read sequencing based whole genome sequencing (WGS) and whole exome sequencing (WES) was performed on a large NIID family (13 affected and 7 unaffected members). Linkage analysis on SNPs extracted from these data revealed only two linked regions: a 3.5 Mb region at 1p36.31-p36.22 (maximum LOD=2.32) and a 58.1 Mb region at 1p22.1-q21.3 (maximum LOD: 4.21). However, no pathogenic SNPs or CNVs were identified in these linked regions.
GGC repeat expansions in NOTCH2NLC
Nanopore-based sequencing was processed on 13 affected and 4 unaffected members from 8 families (another affected member was sequenced by Pacbio long read sequencing platform.). Long-read data revealed disease associated GGC repeat expansions in the 5′ UTR of NOTCH2NLC gene mapping to 58.1 Mb linked region (Figure 1). These repeat expansions were also identified in all 40 sporadic NIID cases tested by RP-PCR.
Cas-9 mediated target sequencing on nanopore platform was employed to achieve higher read coverage on NOTCH2NLC repeat (100 X-1,795 X). These consensus sequences agreed well with previous findings on GGC repeat expansions. Moreover, {(GGA)n (GGC)n}n repeats were identified as a potential genetic marker for weakness-dominant phenotype (Figure 2).
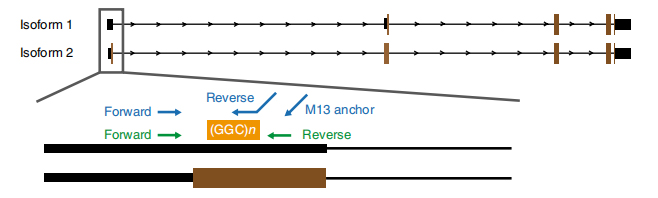
Figure 1. Disease associated repeat expansion identified on exon 1 of NOTCH2NLC isoforms.
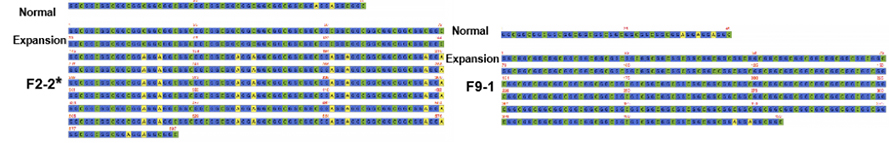
Figure 2. Consensus sequences of NPTCH2NLC repeat in NIID patients with(*) or without weakness-dominant phenotype
NOTCH2NL genes are human-specific genes, which are believed to play vital role in human brain evolution and neurological diseases. However, three NOTCH2 related genes (NOTCH2NLA, NOTCH2NLB and NOTCH2NLC) with >99.1% sequence identity were not resolved until the latest human genome assembly. Synthesis-free and long-read sequencing on nanopore platform have shown notable advantages in resolving regions of high similarity and (GGC)n repeats with 100% GC-rich.
GGC repeat expansions in NOTCH2NLC
Transcriptome sequencing was processed on 2 affected and 2 unaffected members. Normalized read depth was calculated on sense and antisense strands in upstream of first exons of NOTCH2NL paralogs. Abnormal anti-sense transcripts were found only in affected cases, which sit in the beginning or inside the repeat expansion region (Purple peaks in F1-14 and F1-16 in Figure 3.). In addition, 54 DEGs were identified and all were enriched in GO and MPO terms related to neuronal functions.
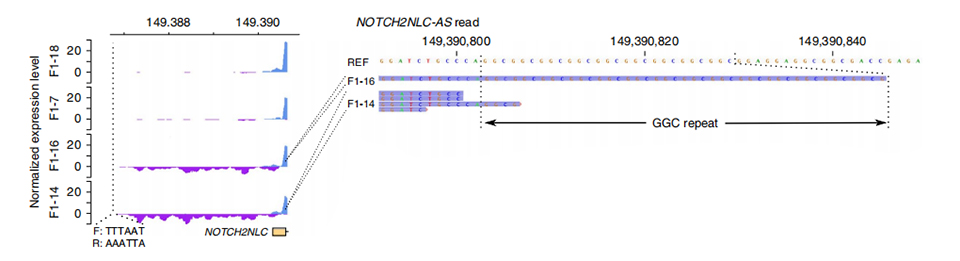
Figure 3. Normalized read depth on upstream of the first exon of NOTCH2NLC in unaffected (above) and affected (below) cases.
Technology
Oxford Nanopore Teghnologies (ONT)
Nanopore sequencing distinguishes itself from other sequencing platforms, in that the nucleotides are read directly without DNA synthesis process. As a single strand DNA passes through a nano-sized protein pore (nanopore), different nucleotides generate different ionic currents, which can be captured and transferred into sequence of bases. ONT sequencing platform itself doesn’t show apparent technical limit on the length of DNA reading. Therefore, Ultra-long reads (ULRs) are available for genome assembly of high quality. Moreover, these extremely long reads, which are long enough to cross complex sequence features or structural variation, help overcome the limitations of short-read sequencing here.

Nanopore sequencing
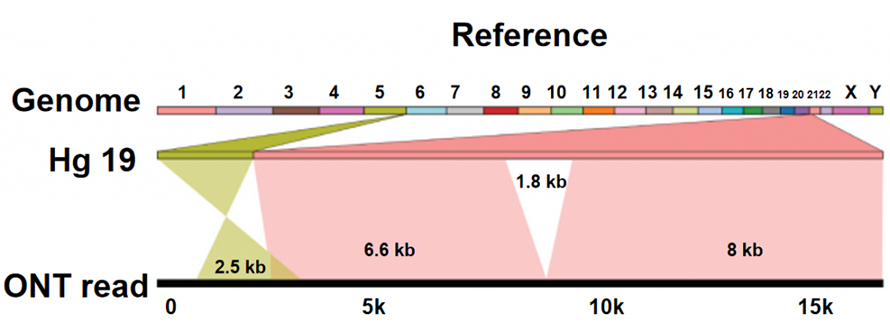
Structure variation (SV) identification
Synthesis-free sequencing largely preserved DNA methylation information on template. Methylated A, T, C and G generate distinct ionic currents from un-methylated ones, which can be read directly by the platform. Nanopore sequencing empowers whole-genome profiling of both 5mC and 6mA at single-nucleotide resolution.
Reference
Jun Sone, et. al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nature Genetics (2019)
Tech and Highlights aims at sharing most recent successful application of different high-throughput sequencing technologies in various reseach arena as well as brilliant ideas in experimental design and data mining .
Post time: Jan-06-2022

