Ke Kaʻina Metagenomic -NGS
Pono lawelawe
● Hoʻokaʻawale a mahiʻai ʻole no ka hoʻopili ʻana i ke kaiāulu microbial
● Hoʻoholo kiʻekiʻe i ka ʻike ʻana i nā ʻano haʻahaʻa haʻahaʻa i nā laʻana kaiapuni
● Hoʻohui ka manaʻo o "meta-" i nā hiʻohiʻona olaola a pau ma ka pae hana, ka pae ʻano a me ka pae gene, e hōʻike ana i kahi hiʻohiʻona ikaika e pili kokoke ana i ka ʻoiaʻiʻo.
● Hoʻoulu ʻo BMK i ka ʻike nui ma nā ʻano laʻana like ʻole me ka ʻoi aku o 10,000 mau laʻana i hana ʻia.
Nā kikoʻī lawelawe
| Papahana | Ke kaʻina ʻana | Manaʻo ʻikepili | Ka manawa huli |
| ʻO Illumina NovaSeq Platform | PE150 | 6 G/10 G/20 G | 45 mau lā hana |
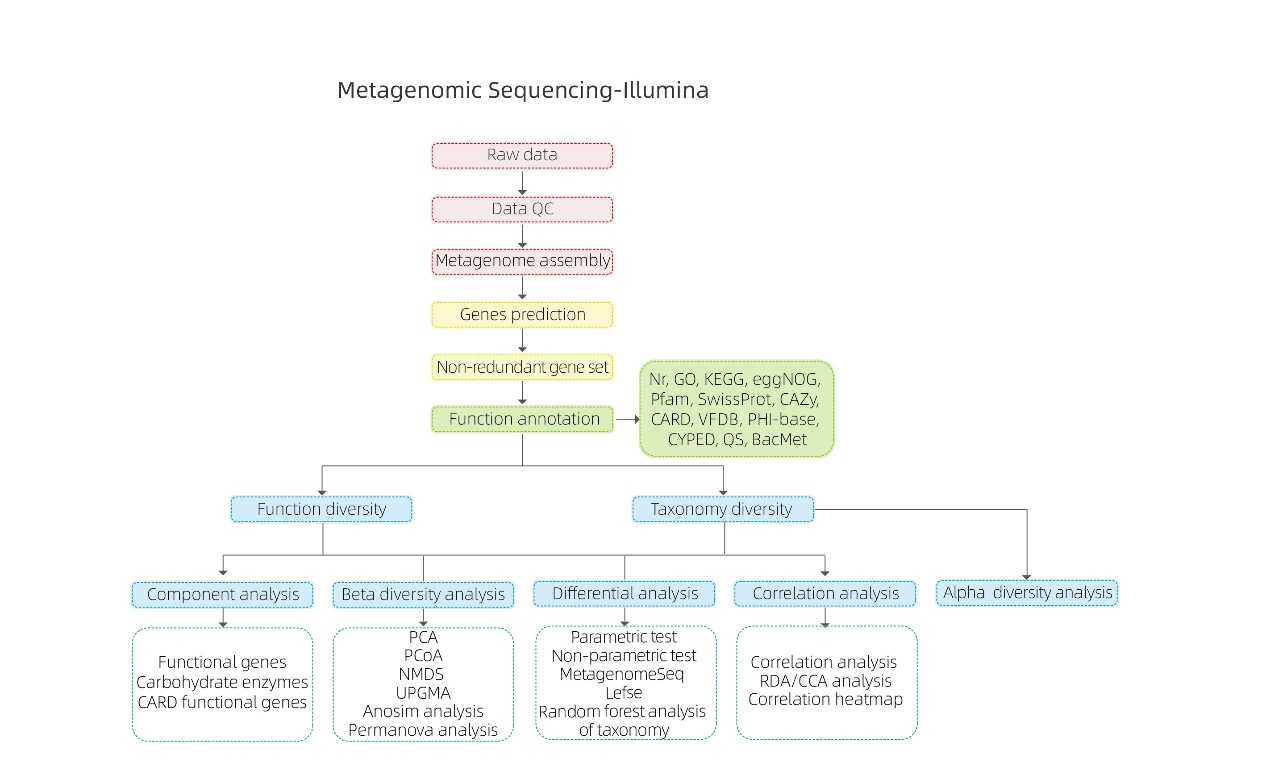
Nā loiloi bioinformatics
● Ka mana o ka maikaʻi o ka ʻikepili maka
● Hui Metagenome
● ʻAʻole-redundant gene set and annotation
● Ka nānā 'ana i nā 'ano like 'ole
● Ke kālailai ʻokoʻa hana genetic
● Ka nānā ʻana ma waena o ka hui
● Kānāwai hui kūʻē i nā kumu hoʻokolohua
Nā Koina Laʻana a me ka Hoʻouna ʻana
Nā Koina Laʻana:
No ka meaNā unuhi DNA:
| ʻAno Laʻana | Ka nui | Hoʻopaʻa | Maemae |
| Nā unuhi DNA | > 30 ng | > 1 ng/μl | OD260/280= 1.6-2.5 |
No nā laʻana kaiapuni:
| ʻAno laʻana | Manaʻo ʻia ke kaʻina hana laʻana |
| lepo | Ka nui o ka laʻana: approx.5 g;Pono e wehe ʻia nā mea maloʻo i koe mai ka ʻili;E wili i nā ʻāpana nui a hele i ka kānana 2 mm;ʻO nā laʻana Aliquot i loko o ka sterile EP-tube a i ʻole cyrotube no ka mālama ʻana. |
| Feces | Ka nui o ka laʻana: approx.5 g;E hōʻiliʻili a hāʻawi i nā laʻana i loko o ka sterile EP-tube a i ʻole cryotube no ka mālama ʻana. |
| ʻO loko o ka ʻōpū | Pono e hana ʻia nā laʻana ma lalo o ke kūlana aseptic.Holoi i ka ʻiʻo i hōʻiliʻili ʻia me ka PBS;Centrifuge i ka PBS a ohi i ka precipitant ma EP-tubes. |
| Sludge | Ka nui o ka laʻana: approx.5 g;E hōʻiliʻili a hāʻawi i kahi laʻana o ka lepo i loko o ka EP-tube sterile a i ʻole cryotube no ka mālama ʻana |
| Ke kino wai | No ka laʻana me ka liʻiliʻi o ka microbial, e like me ka wai paipu, ka wai punawai, a me nā mea ʻē aʻe, E hōʻiliʻili i ka liʻiliʻi o 1 L wai a hele i ka kānana 0.22 μm e hoʻonui i ka microbial ma ka membrane.E mālama i ka membrane i loko o ka paipu sterile. |
| ʻili | E ʻoki pono i ka ʻili o ka ʻili me ka pulupulu maʻemaʻe a i ʻole ka lāʻau lapaʻau a waiho i loko o ka paipu sterile. |
Manaʻo ʻia ka hāʻawi laʻana
E hoʻokuʻu i nā laʻana i loko o ka hau hau no 3-4 mau hola a mālama i loko o ka wai nitrogen a i ʻole -80 degere a hiki i ka mālama lōʻihi.Pono ka hoʻouna laʻana me ka hau maloʻo.
Holo Hana Hana

Hāʻawi laʻana

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili

Nā lawelawe ma hope o ke kūʻai aku
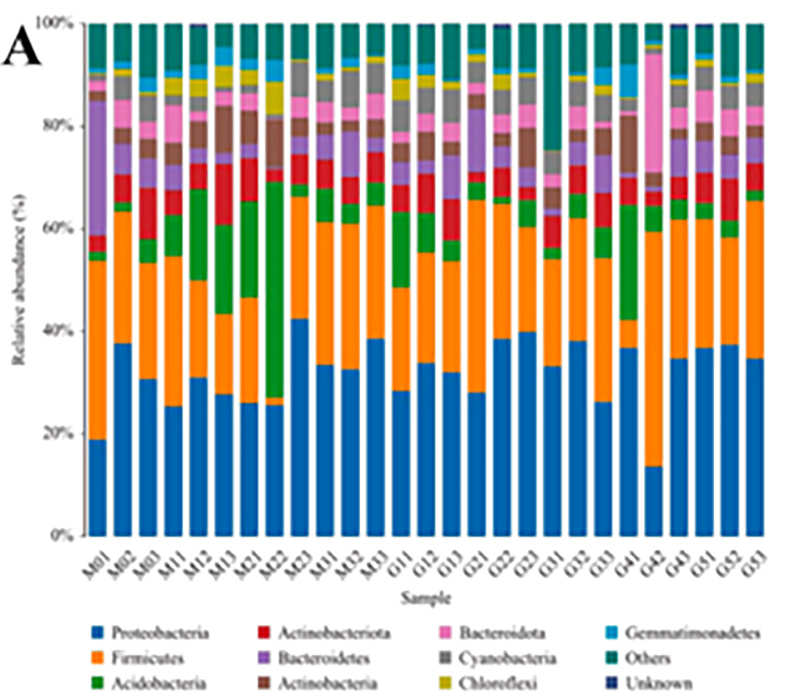
1.Histogram: Hoʻolaha ʻano ʻano
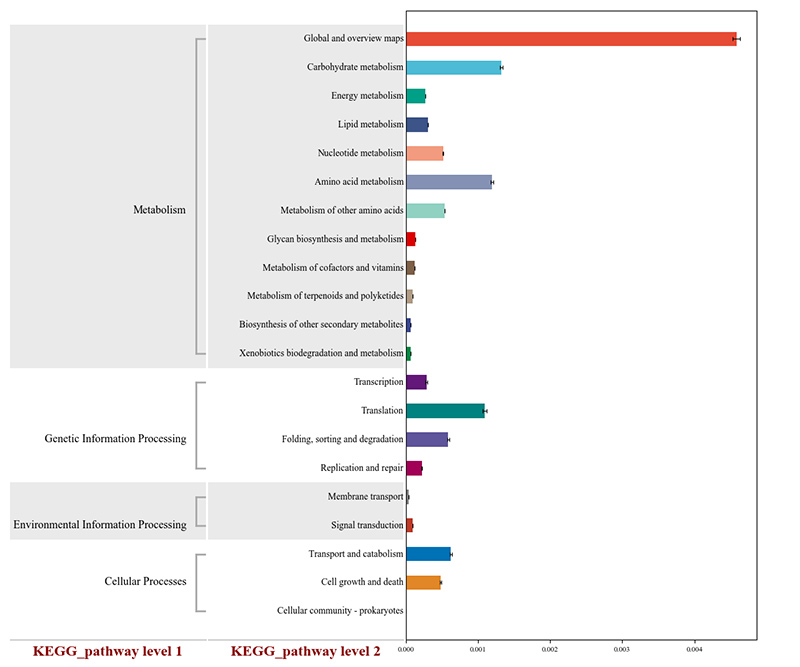
2.Functional genes annotated to KEGG metabolic pathways
3. Heat map: Nā hana like ʻole e pili ana i ka nui o nā gene pili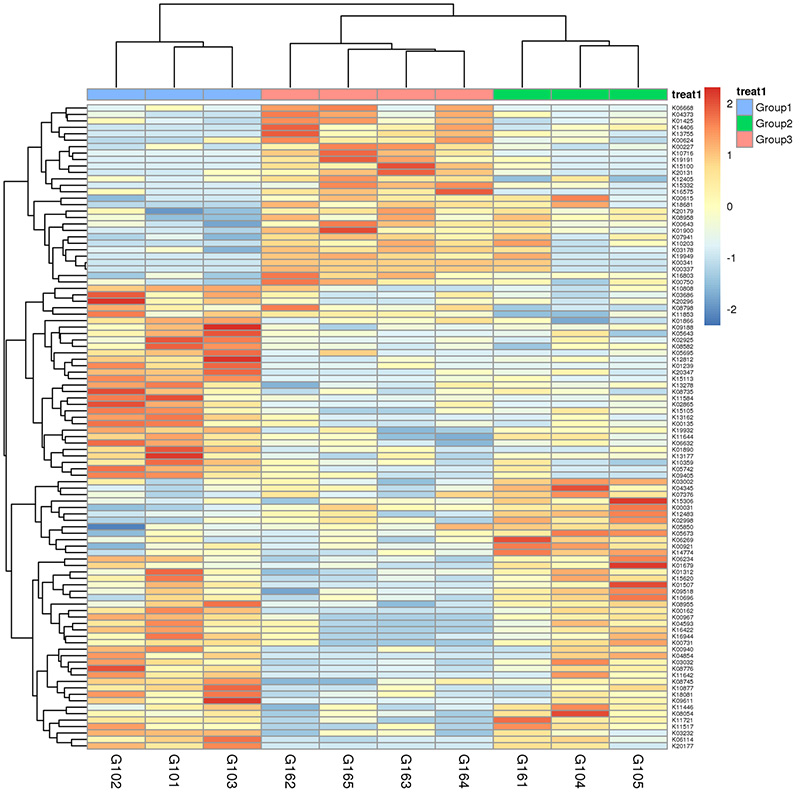 4.Circos o CARD antibiotic kū'ē genes
4.Circos o CARD antibiotic kū'ē genes
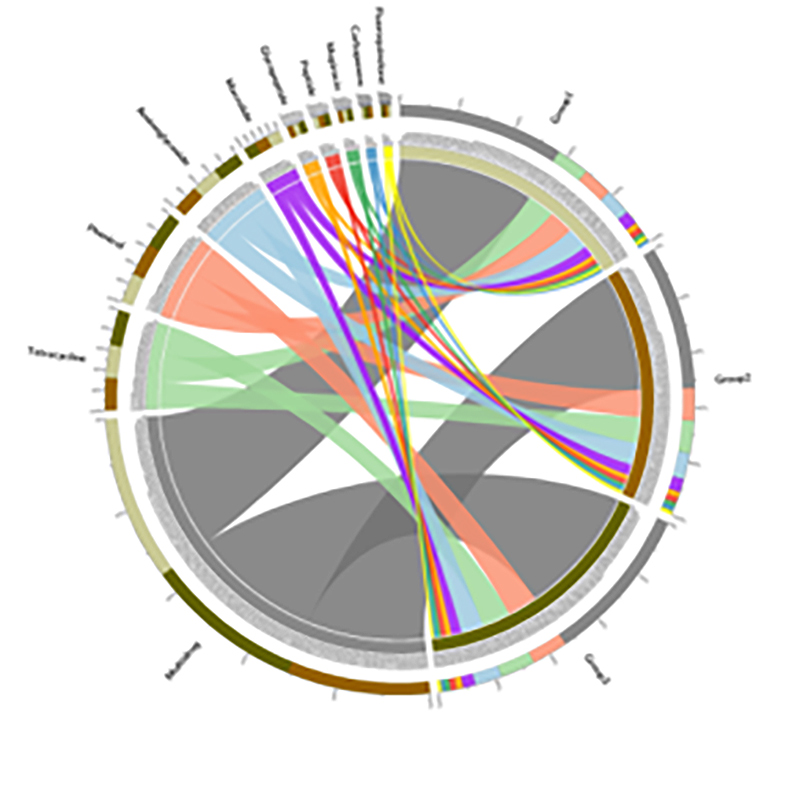
Hihia BMK
ʻO ka laha ʻana o nā genes pale antibiotic a me nā maʻi bacterial ma ka hoʻomau ʻana o ka lepo-mangrove root continuum
Paʻi ʻia:Nūpepa o nā Mea Pilikino, 2021
Hoʻolālā kaʻina hana:
Mea waiwai: Nā ʻāpana DNA o nā ʻāpana ʻehā o nā kumu lāʻau mangrove e pili ana: lepo kanu ʻole, rhizosphere, episphere a me nā keʻena endosphere.
Papahana: Illumina HiSeq 2500
Nā pahuhopu: Metagenome
16S rRNA gene V3-V4 māhele
Nā hopena koʻikoʻi
Ua hoʻoponopono ʻia ka metagenomic sequencing a me ka metabarcoding profiling ma ka ʻaʻa lepo o nā kumu lāʻau mangrove i mea e aʻo ai i ka hoʻolaha ʻana o nā genes resistance antibiotic (ARGs) mai ka lepo i nā mea kanu.Ua hōʻike ʻia ka ʻikepili Metagenomic he 91.4% o nā genes resistance antibiotic i ʻike maʻamau i loko o nā keʻena lepo ʻehā i ʻōlelo ʻia ma luna, kahi i hōʻike ʻia i ke ʻano mau.ʻO 16S rRNA amplicon sequencing i hana i 29,285 sequencing, e hōʻike ana i 346 mau ʻano.I ka hui pū ʻana me ka hoʻopili ʻana i nā ʻano mea ma o ka amplicon sequencing, ua ʻike ʻia kēia hoʻolaha ʻana he kūʻokoʻa i ka microbiota pili i ke aʻa, akā naʻe, hiki ke hoʻomaʻamaʻa ʻia e ka mobile o nā mea genetic.Ua ʻike kēia haʻawina i ka kahe ʻana o nā ARG a me nā pathogens mai ka lepo i loko o nā mea kanu ma o ka hoʻomau ʻana o ka lepo-aʻa.
Kuhikuhi
Wang, C. , Hu, R. , Strong, PJ , Zhuang, W. , & Shu, L. .(2020).ʻO ka laha ʻana o nā genes kūpaʻa antibiotic a me nā maʻi bacterial ma ka lepo-aʻa kumu mangrove.Nūpepa o nā mea pōʻino, 408, 124985.