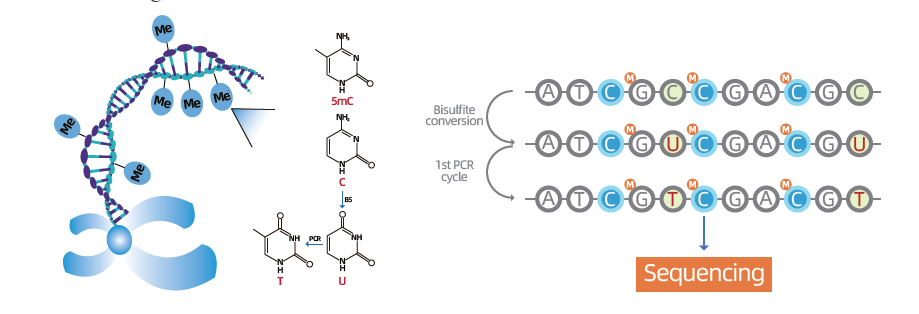
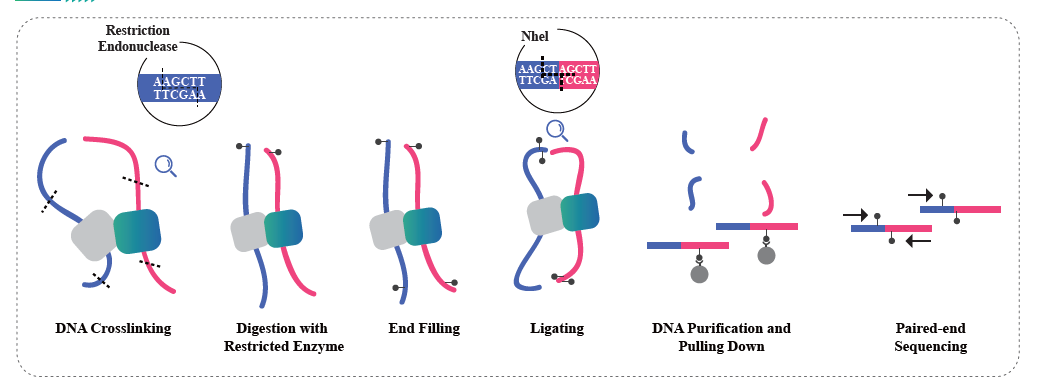
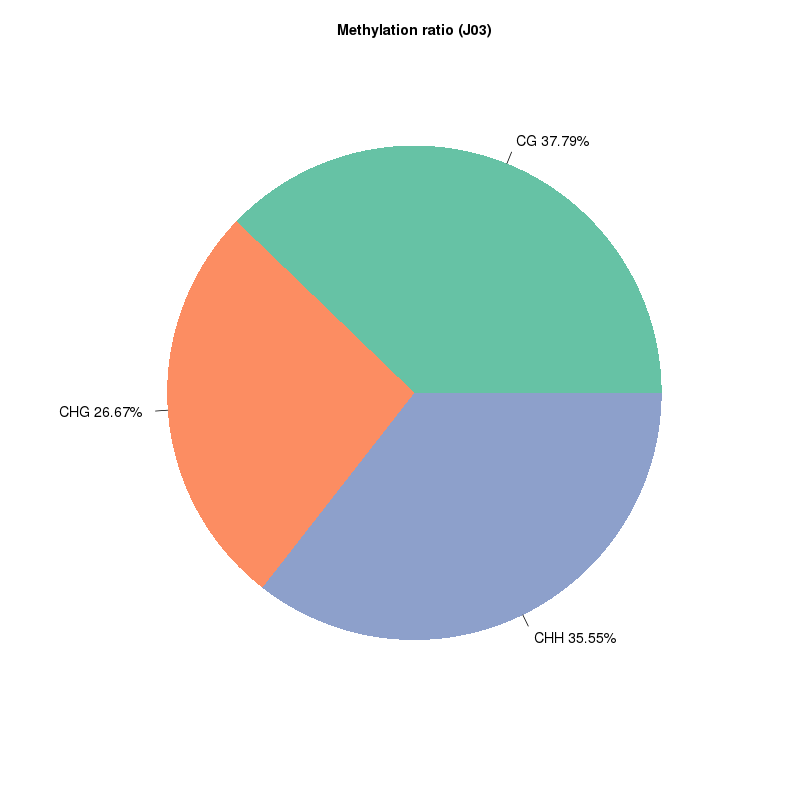Whole genome bisulfite sequencing(WGBS)
Service Features
● Requires a reference genome.
● Lambda DNA is added to monitor bisulfite conversion efficiency.
● Sequencing on Illumina NovaSeq.
Service Advantages
● Gold standard for DNA methylation research: this mature methylation conversion processing technology has high accuracy and good reproducibility.
● Wide coverage and single-base resolution: detection of methylation sites at the genome-wide level.
● Complete platform: provide one-stop excellent service from sample processing, library construction, sequencing to bioinformatics analysis.
● Extensive Expertise: with WGBS sequencing projects successfully completed across a diverse range of species, BMKGENE brings over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
● Possibility to join with transcriptomics analysis: allowing for the integrated analysis of WGBS with other omics data such as RNA-seq.
Sample Specifications
|
Library |
Sequencing Strategy |
Recommended data output |
Quality control |
|
Bisulfite treated |
Illumina PE150 |
30x depth |
Q30≥85% Bisulfite conversion >99% |
Sample Requirements
|
Concentration (ng/µL) |
Total amount (µg) |
Additional requirements |
|
|
Genomic DNA |
≥30 |
≥3 |
Limited degradation or contamination |
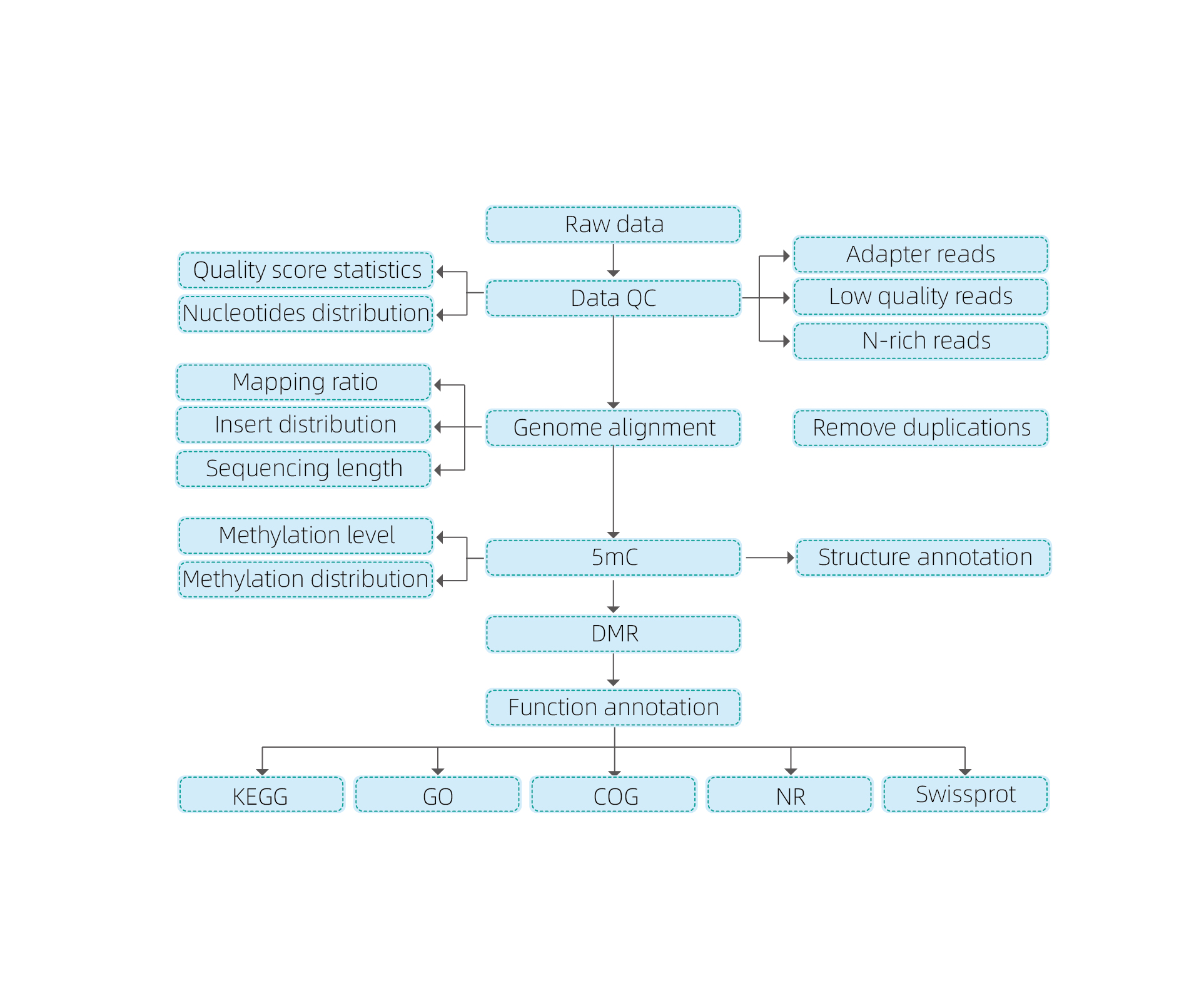
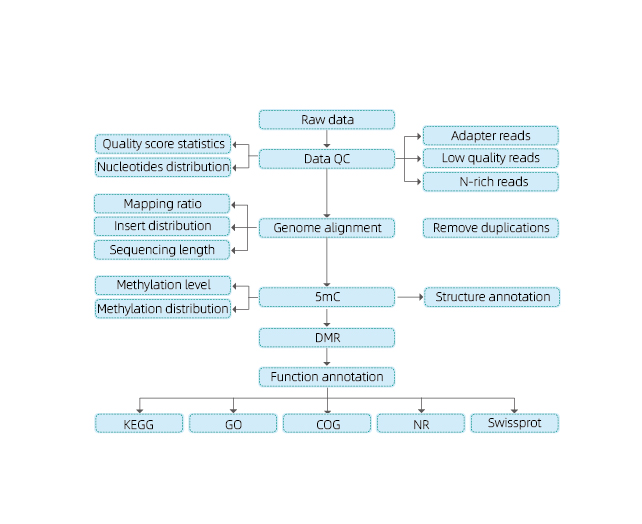
Service Work Flow

Sample delivery

DNA extraction

Library construction

Sequencing

Data analysis
Data delivery
Includes the following analysis:
● Raw sequencing quality control;
● Mapping to reference genome;
● Detection of 5mC methylated bases;
● Analysis of methylation distribution and annotation;
● Analysis of Differentially Methylated Regions (DMRs);
● Functional annotation of genes associated to DMRs.
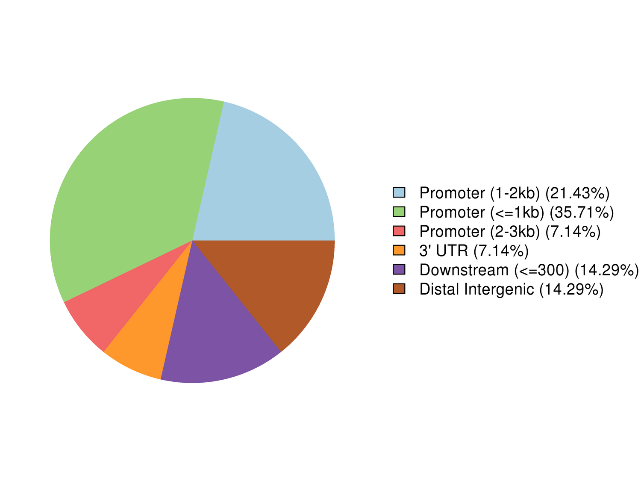
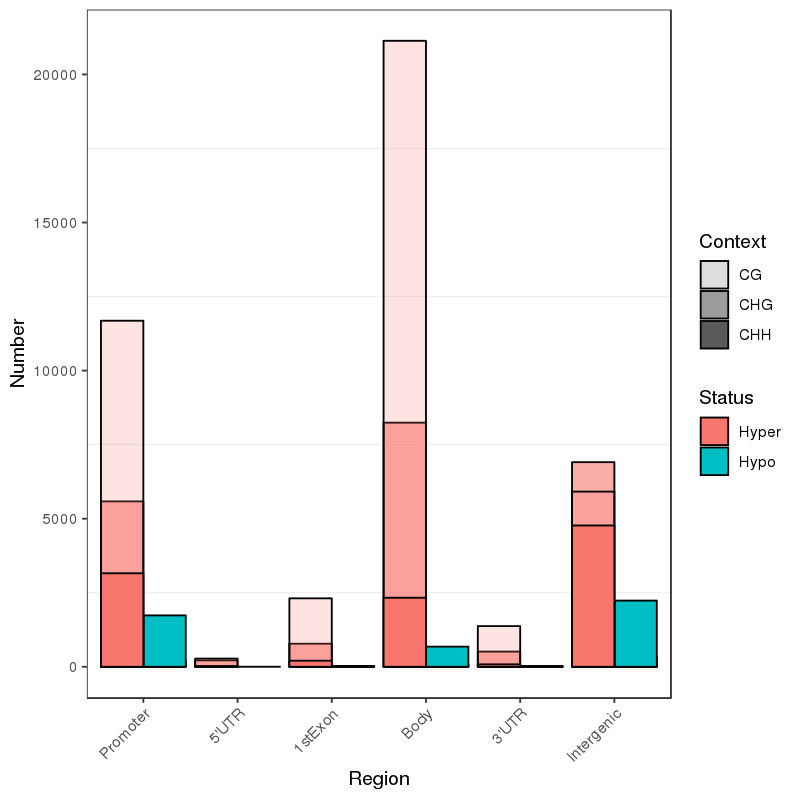
5mC methylation detection: types of methylated sites
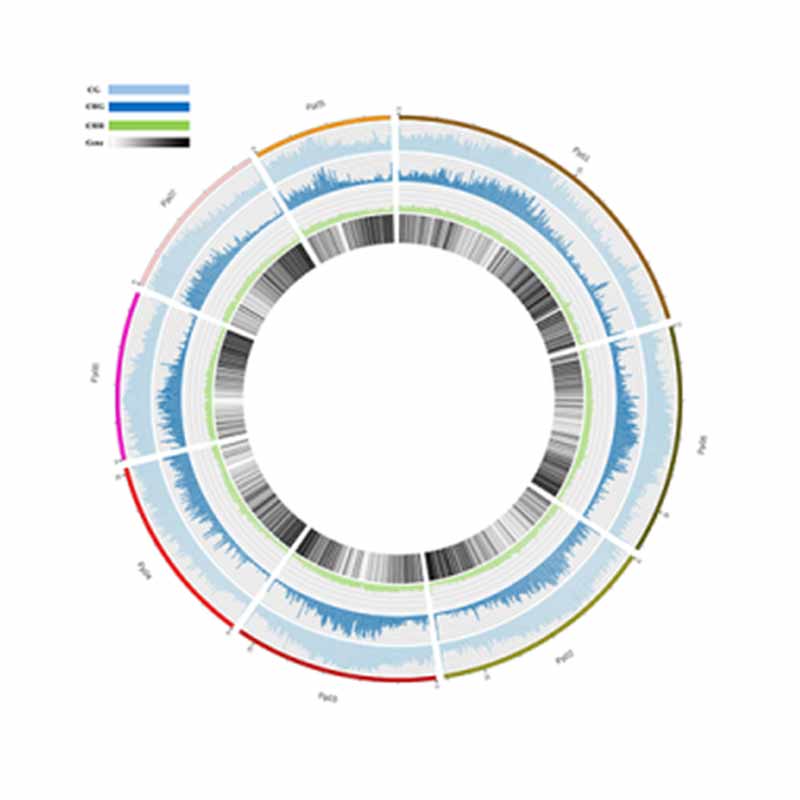
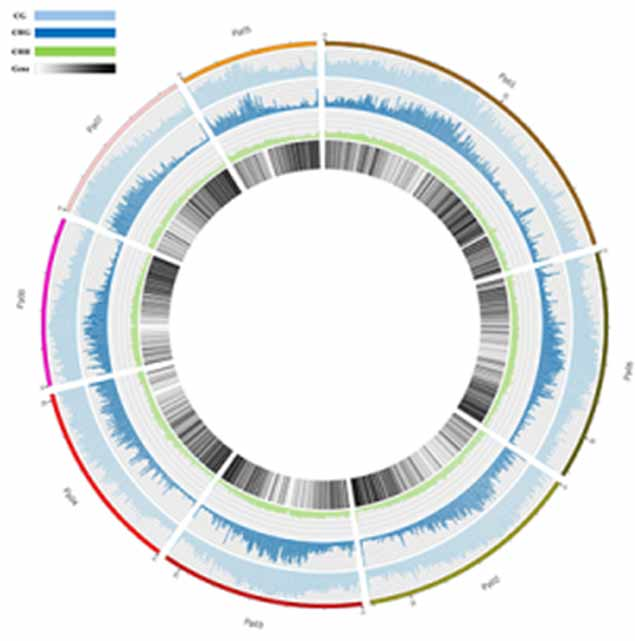
Methylation map. 5mC methylation genome-wide distribution
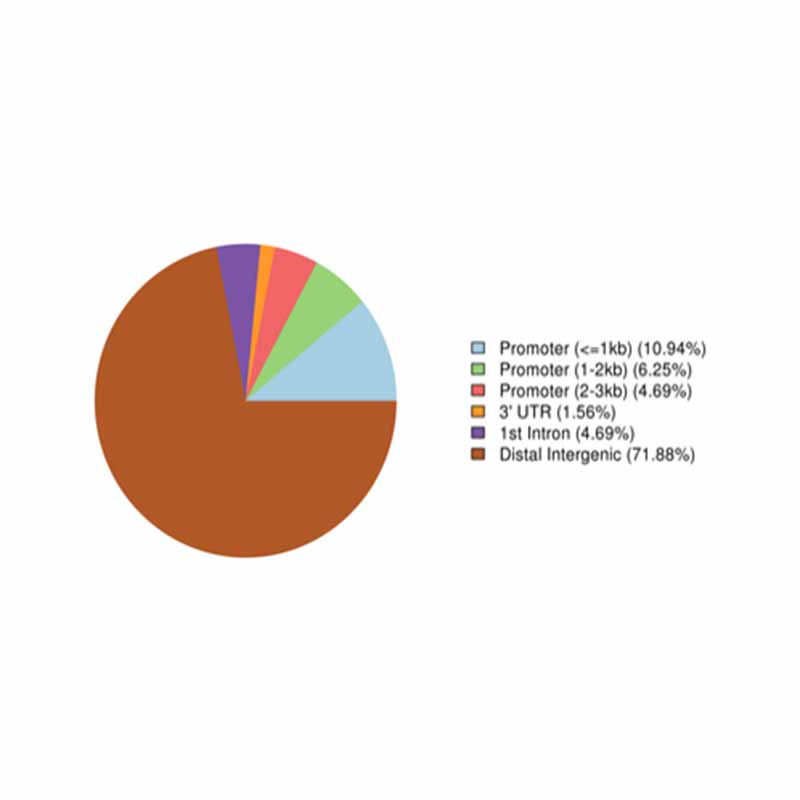
Annotation of highly methylated regions
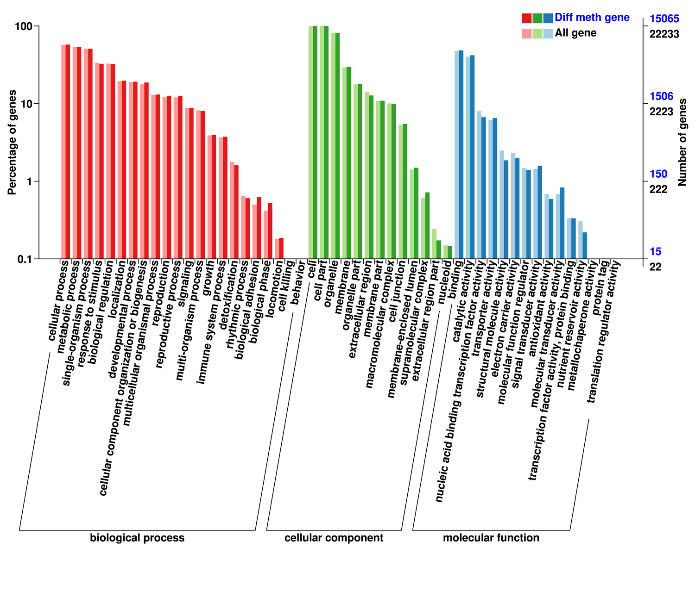
Differentially Methylated Regions: associated genes
Differentially Methylated Regions: annotation of associated genes (Gene Ontology)
Explore the research advancements facilitated by BMKGene’s whole genome bisulfite sequencing services through a curated collection of publications.
Fan, Y. et al. (2020) ‘Analysis of DNA methylation profiles during sheep skeletal muscle development using whole-genome bisulfite sequencing’, BMC Genomics, 21(1), pp. 1–15. doi: 10.1186/S12864-020-6751-5.
Zhao, X. et al. (2022) ‘Novel deoxyribonucleic acid methylation perturbations in workers exposed to vinyl chloride’, Toxicology and Industrial Health, 38(7), pp. 377–388. doi: 10.1177/07482337221098600
Zuo, J. et al. (2020) ‘Relationships between genome methylation, levels of non-coding RNAs, mRNAs and metabolites in ripening tomato fruit’, The Plant Journal, 103(3), pp. 980–994. doi: 10.1111/TPJ.14778.