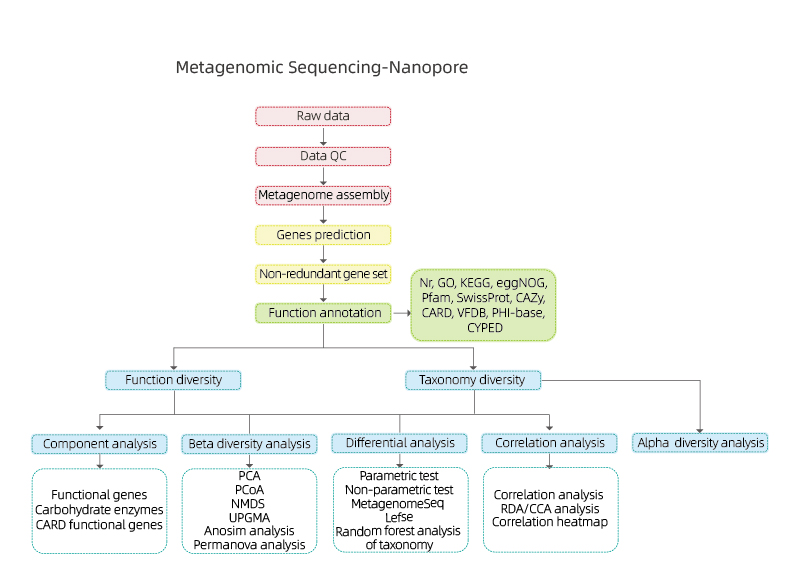
Metagenomic Sequencing-Nanopore
Service Features
● Long read sequencing on Nanopore P48 (8-10Kb library)
● Error-correction: sequencing on Illumina NovaSeq (PE150 library)
Service Advantages
● High-quality metagenome assembly: with less contigs that have higher continuity.
● Higher accuracy: of species identification and functional gene prediction.
● Improved assembly: facilitates bacterial genome isolation and comparative metagenome analysis.
Comprehensive bioinformatics analysis: focused not only on taxonomic diversity but also on the functional diversity of the community.
● Extensive experience: with a track record of successfully closing multiple metagenomics projects in various research domains and processing over 10,000 samples, our team brings a wealth of experience to every project.
Service Specifications
|
Sequencing platform |
Sequencing Strategy |
Data recommended |
|
Illumina NovaSeq |
PE150 |
6-20Gb |
|
Nanopore P48 |
8-10kb |
6-20Gb |
Sample Requirements
|
Concentration (ng/µL) |
Total amount (µg) |
Volume (µL) |
OD260/280 |
OD260/230 |
|
|
Illumina PE150 Library |
≥1 |
≥0.03 |
≥20 |
1.6-2.5 |
- |
|
Nanopore 8-10Kb library |
≥40 |
≥2 |
≥20 |
1.7-2.2 |
≥1.0 |
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
Includes the following analysis:
● Sequencing data quality control
● Metagenome assembly and gene prediction
● Gene annotation
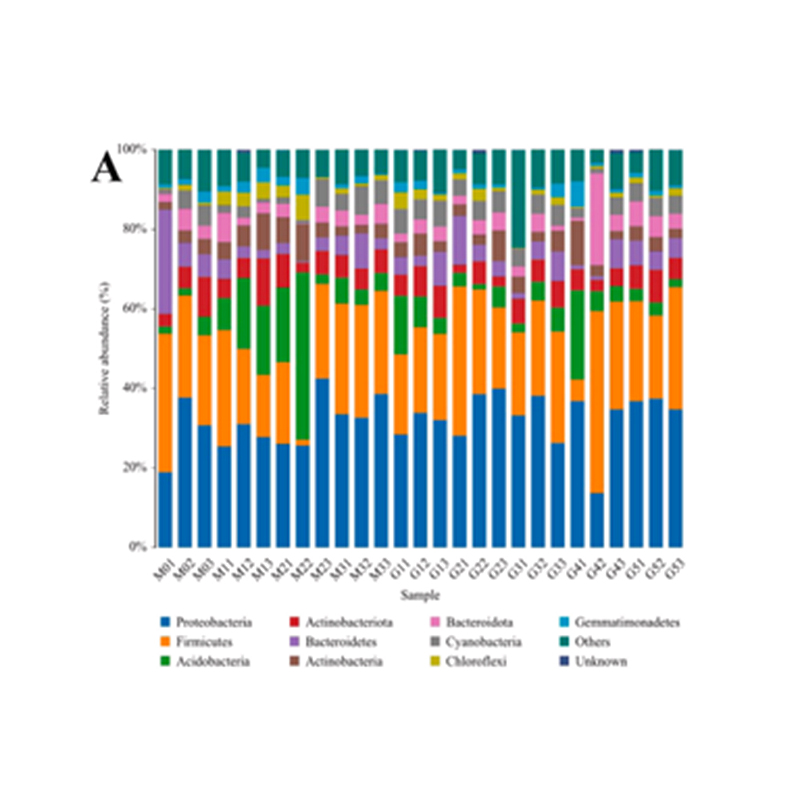
● Taxonomic alpha diversity analysis
● Functional analysis of the community: biological function, metabolic, antibiotic resistance
● Analysis on both functional and taxonomic diversity:
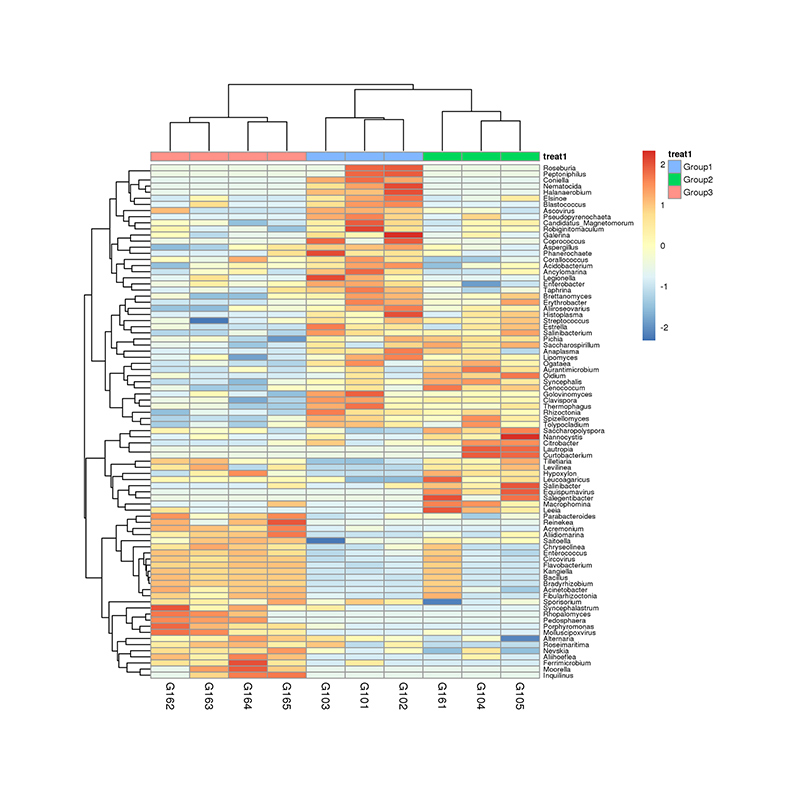
Beta diversity analysis
Inter-group analysis
Correlation analysis: between environmental factors and OUT composition and diversity
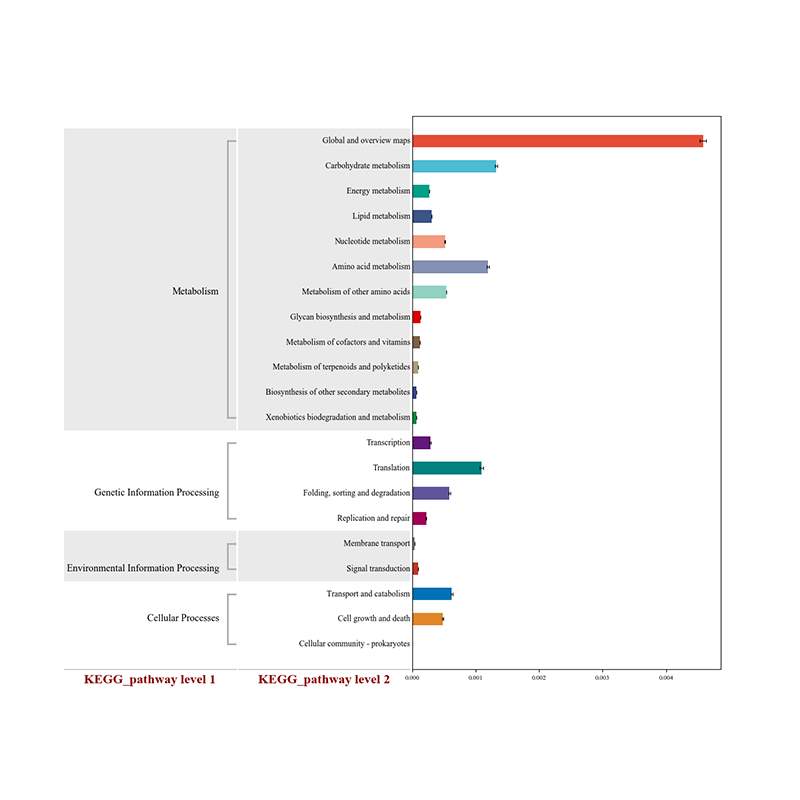
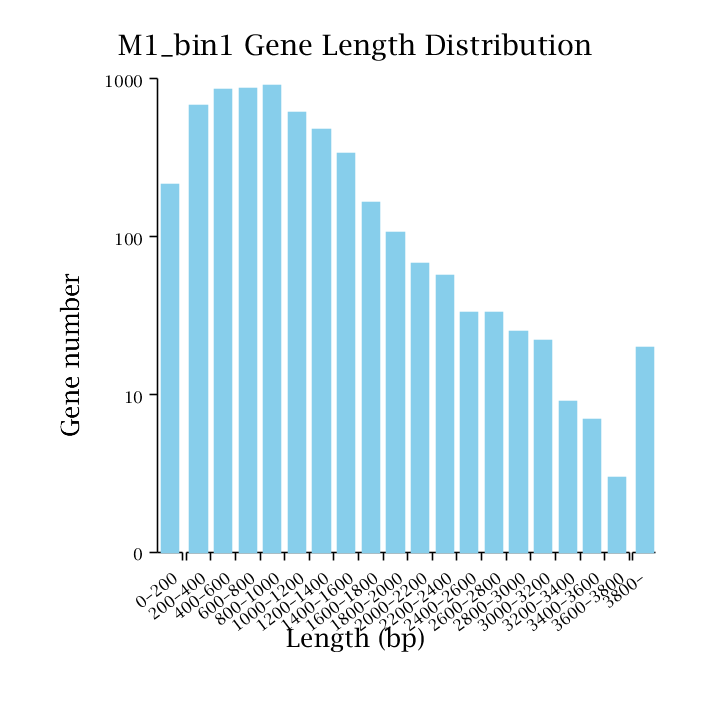
Non-redundant gene set distribution:
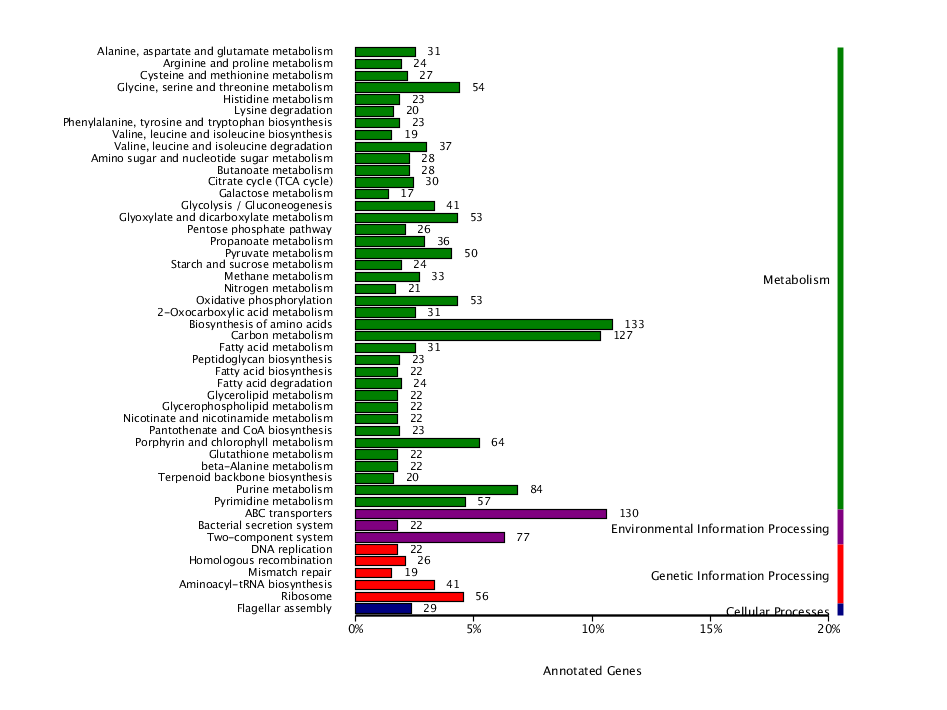
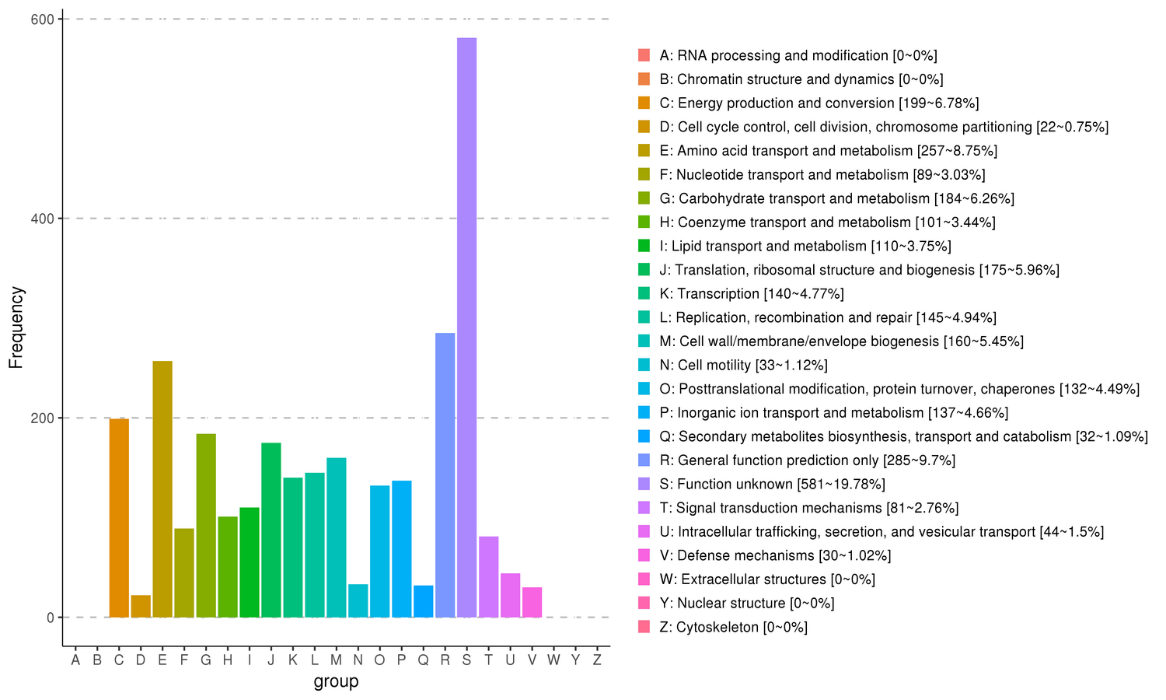
Gene annotation: eggNOG
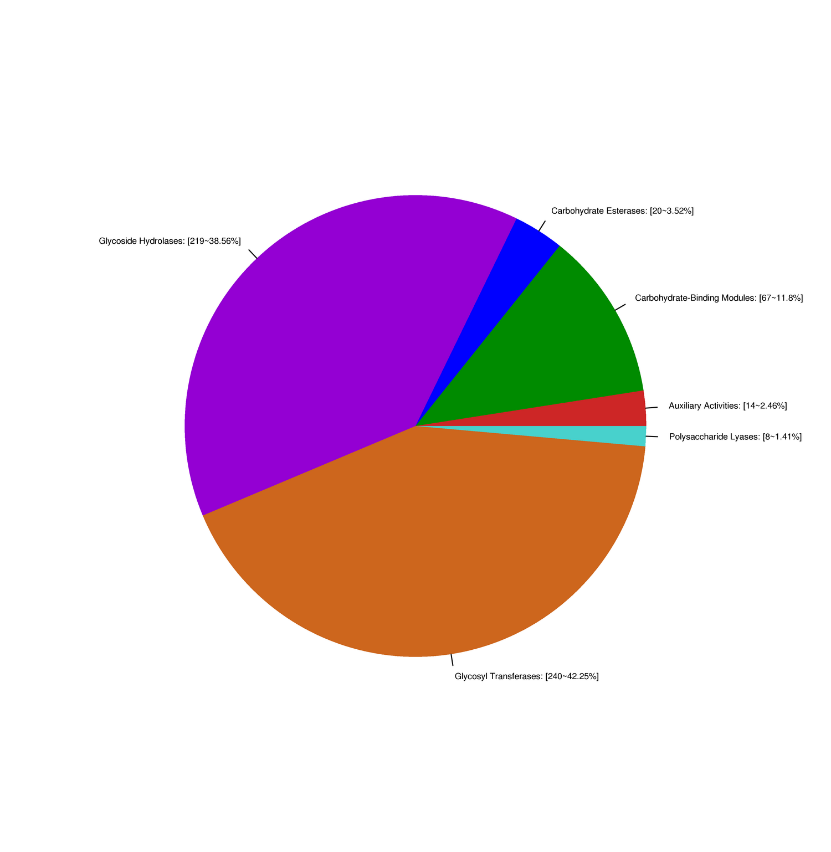
Gene annotation: CAZY carbohydrates
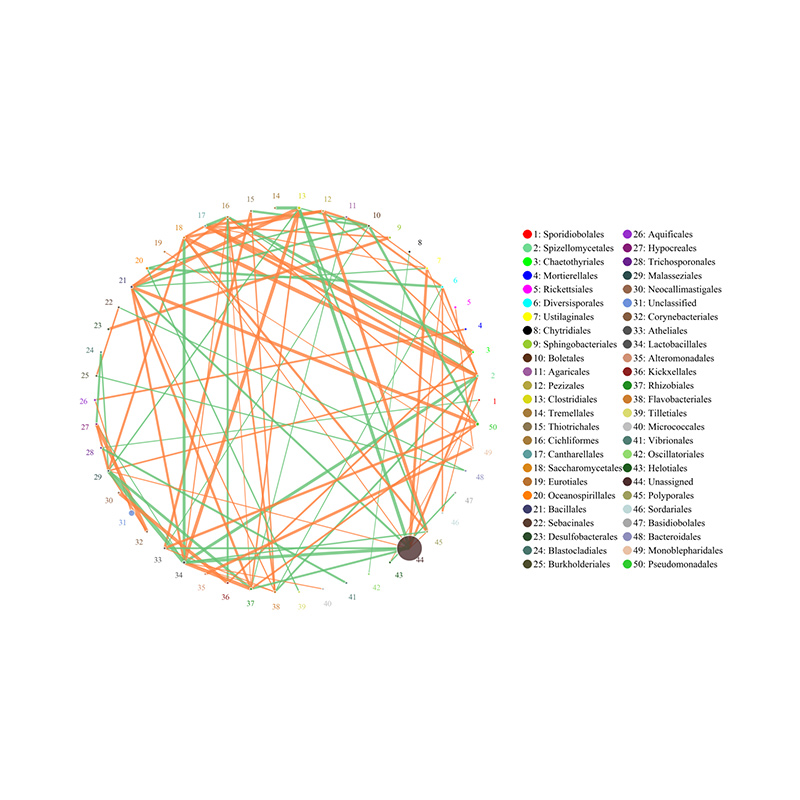
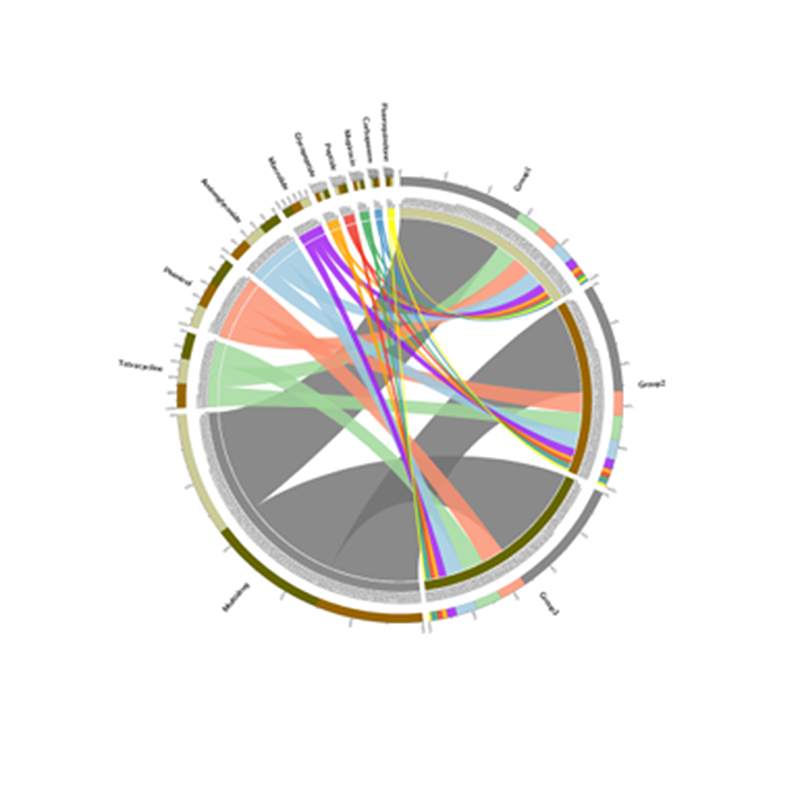
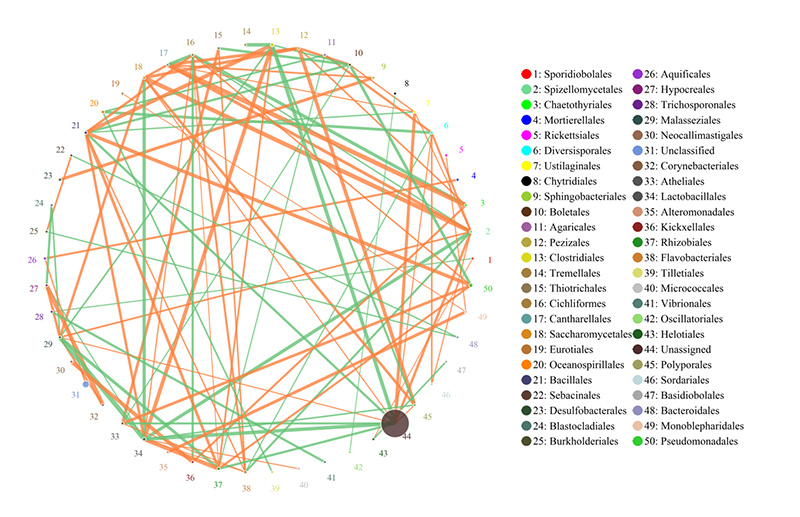
Species correlation network:
Explore the advancements facilitated by BMKGene’s metagenomics sequencing service with Nanopore + Illumina with this featured publication:
Jin, H. et al. (2023) ‘A high-quality genome compendium of the human gut microbiome of Inner Mongolians’, Nature Microbiology 2023 8:1, 8(1), pp. 150–161. doi: 10.1038/s41564-022-01270-1.