16S/18S/ITS Amplicon Sequencing-NGS
Service Features
● Sequencing platform: Illumina NovaSeq.
● Amplification of short regions of 16S, 18S and ITS, among other amplification targets.
● Flexible choices of amplicon.
● Previous project experience with multiple amplification targets.
Service Advantages
● Isolation-free: and rapid identification of microbial composition in environmental samples.
● High resolution: in low-abundant components in environmental samples.
● Widely applicable: to diverse microbial community studies.
● Comprehensive bioinformatic analysis: including the with latest QIIME2 package (quantitative insight into microbial ecology) with diverse analyses in terms of database, annotation, OTU/ASV.
● Extensive Expertise: with thousands of amplicon sequencing projects conducted annually, BMKGENE brings over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
Service Specifications
|
Library |
Sequencing Strategy |
Data recommended |
Quality control |
|
Amplicon |
Illumina PE250 |
50/100K tags (read pairs) |
Q30≥85% |
Service Requirements
|
Concentration (ng/µL) |
Total amount (ng) |
Volume (µL) |
OD260/280 |
|
≥1 |
≥300 |
≥20 |
1.6-2.5 |
● Soil/sludge: 1-2g
● Intestinal content-animal: 0.5-2g
● Intestinal contents-insect: 0.1-0.25g
● Plant surface (enriched sediment): 0.1-0.5g
● Fermentation broth enriched sediment): 0.1-0.5g
● Faeces (large animals): 0.5-2g
● Faeces (mouse): 3-5grains
● Pulmonary alveolar lavage fluid: filter paper
● Vaginal swab: 5-6 swabs
● Skin/genital swab/saliva/oral soft tissue/pharyngeal swab/rectal swab: 2-3 swabs
● Surface microorganisms: filter paper
● Waterbody/air/biofilm: filter paper
● Endophytes: 1-2g
● Dental Plaque: 0.5-1g
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
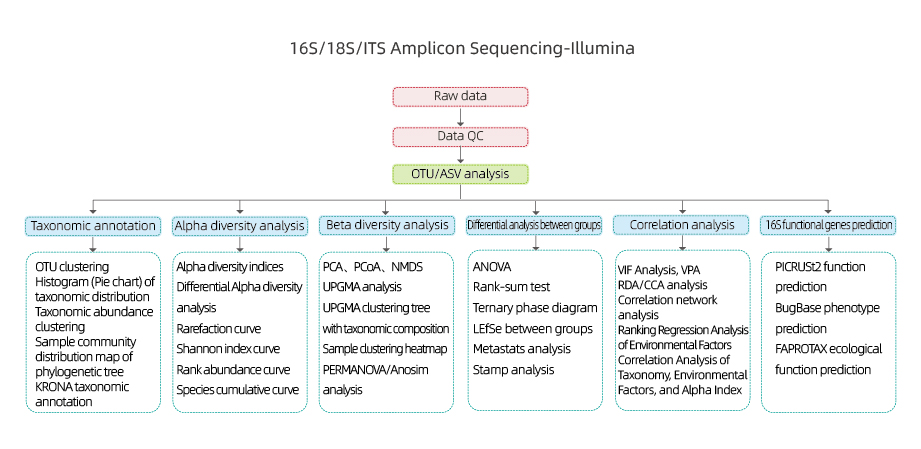
Includes the following analysis:
- Raw data quality control
- OTU clustering /De-noise(ASV)
- OTU annotation
- Alpha diversity analysis: multiple indexes, including Shannon, Simpson and ACE.
- Beta diversity analysis
- Inter-group analysis
- Correlation analysis: between environmental factors and OUT composition and diversity
- 16S functional gene prediction
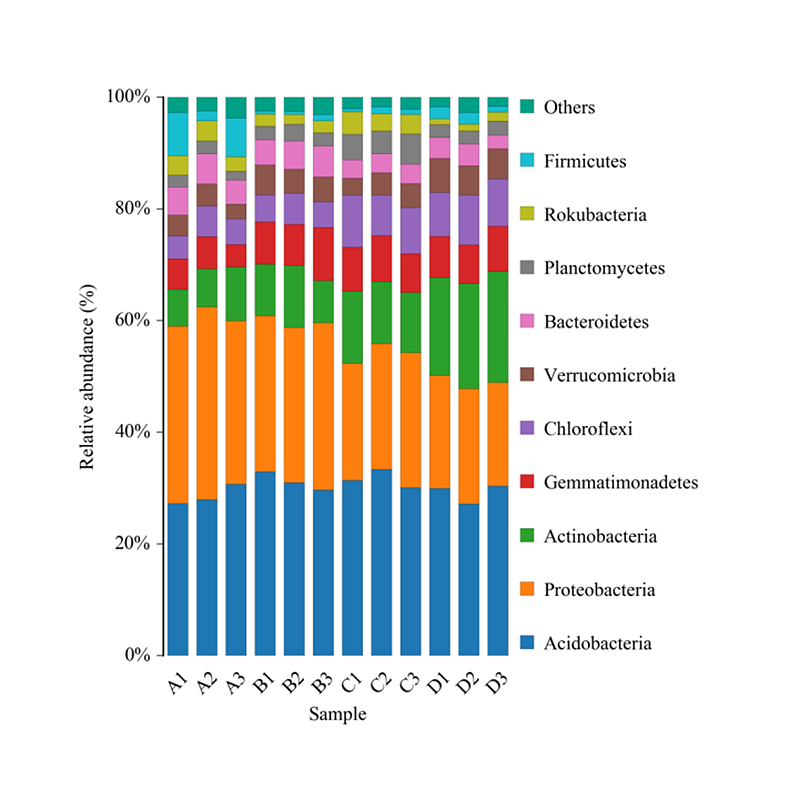
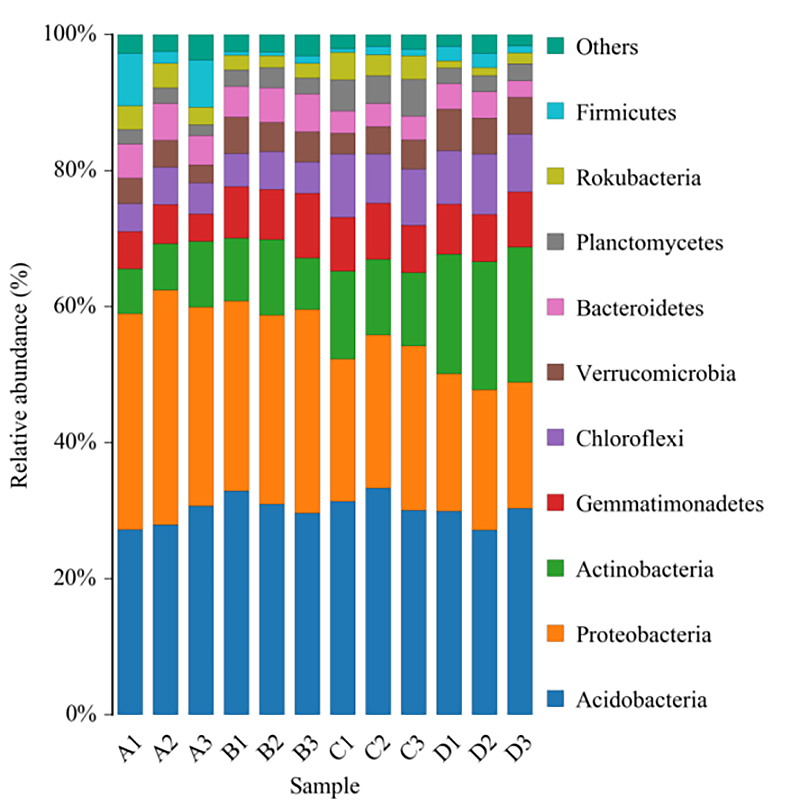
Histogram of taxonomic distribution
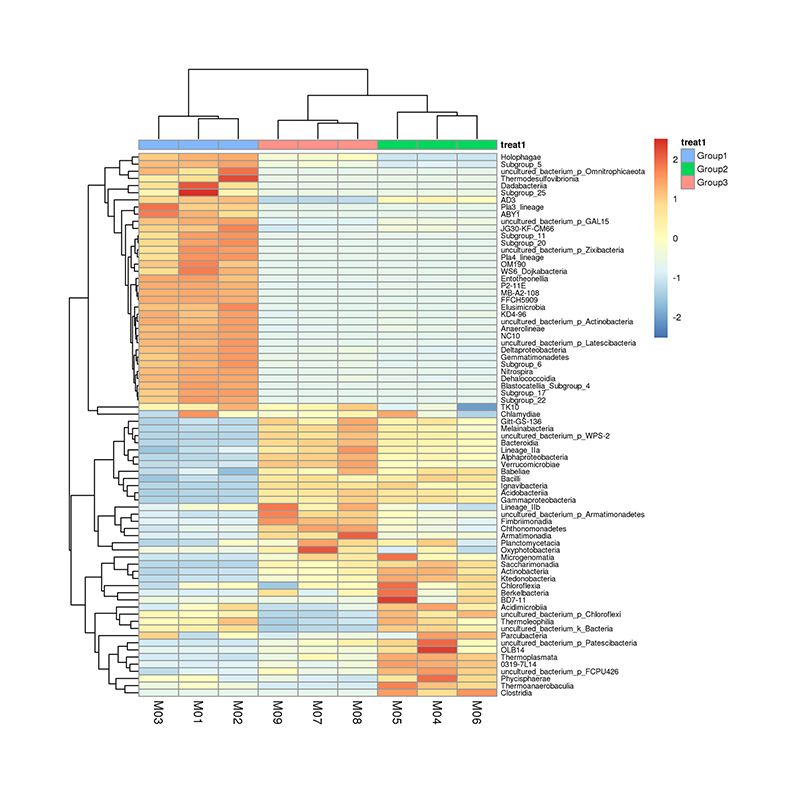
taxonomic abundance clustering heat map
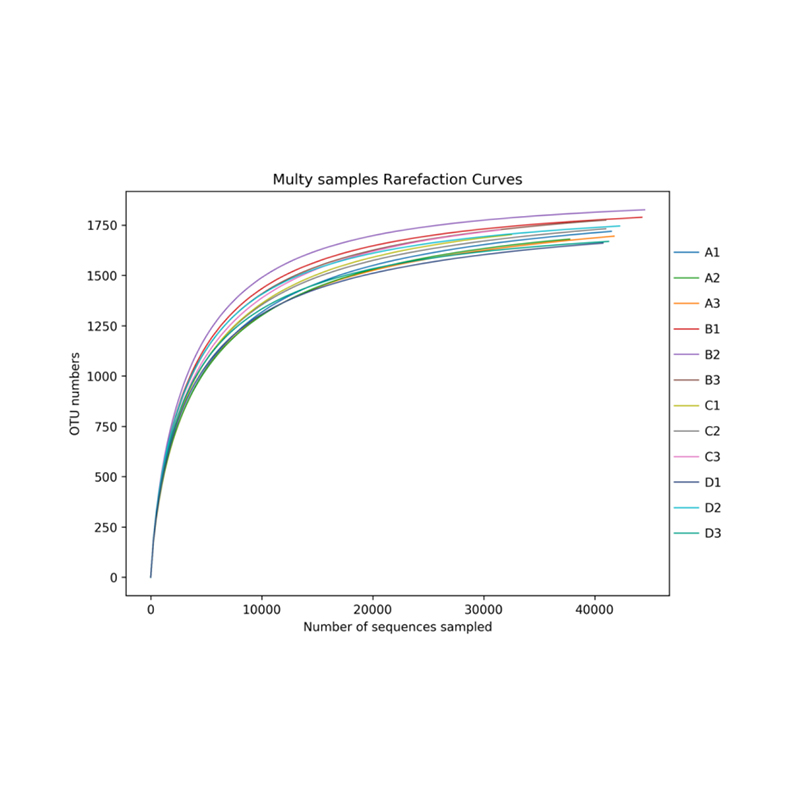
Alpha diversity analysis: rarefaction curve
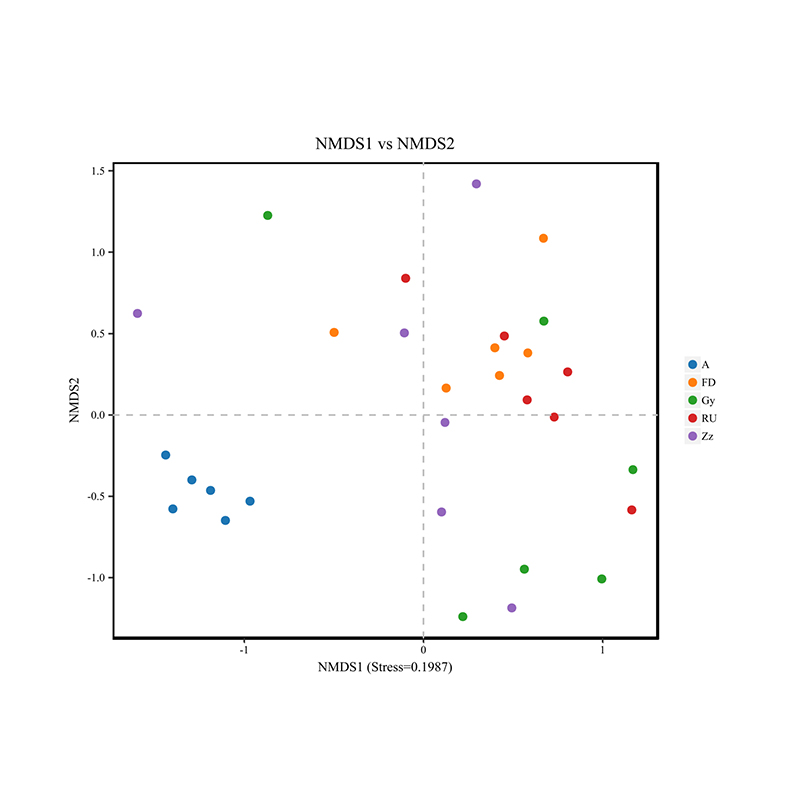
beta diversity analysis: NMDS
Intergroup analysis: LEFSE biomarker discovery
Explore the advancements facilitated by BMKGene’s amplicon sequencing services with Illumina through a curated collection of publications.
Dong, C. et al. (2022) ‘Assembly, Core Microbiota, and Function of the Rhizosphere Soil and Bark Microbiota in Eucommia ulmoides’, Frontiers in Microbiology, 13. doi: 10.3389/FMICB.2022.855317/FULL.
Li, Y. et al. (2023) ‘Synthetic bacterial consortia transplantation for the treatment of Gardnerella vaginalis-induced bacterial vaginosis in mice’, Microbiome, 11(1), pp. 1–14. doi: 10.1186/s40168-023-01497-y
Yang, J., Fu, Y. and Liu, H. (2022) ‘Microbiomes of air dust collected during the ground-based closed bioregenerative life support experiment “Lunar Palace 365”’, Environmental Microbiomes, 17(1), pp. 1–20. doi: 10.1186/S40793-022-00399-0/FIGURES/8.
Yin, S. et al. (2022) ‘Feedstock-dependent abundance of functional genes related to nitrogen transformation controlled nitrogen loss in composting’, Bioresource Technology, 361, p. 127678. doi: 10.1016/J.BIORTECH.2022.127678.