Prokaryotic mRNA sequencing
Features
● RNA sample processing involved rRNA depletion followed by directional RNA library preparation.
● Bioinformatic analysis based on alignment to a reference genome
● Analysis includes gene expression and DEGs but also transcript structure and sRNA analysis
Service Advantages
● Rigorous Quality Control: we implement core control points across all stages, from sample and library preparation to sequencing and bioinformatics. This meticulous monitoring ensures the delivery of consistently high-quality results.
● Strand-specific sequencing data: due to the RNA library preparation being directional, enabling identification of anti-sense transcripts.
● Complete analysis tailored to prokaryotic transcriptomes: the bioinformatic pipeline includes not only analysis of the gene expression but also analysis of the transcript structure, including identification of operons, UTRs and promoters. It also includes analysis of sRNAs, namely annotation and prediction of secondary structure and targets.
● Post-Sales Support: our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Sample Requirements and Delivery
|
Library |
Sequencing strategy |
Data recommended |
Quality Control |
|
Poly A enriched |
Illumina PE150 |
1-2 Gb |
Q30≥85% |
Sample Requirements:
|
Conc.(ng/μl) |
Amount (μg) |
Purity |
Integrity |
|
≥ 50 |
≥ 1 |
OD260/280=1.7-2.5 OD260/230=0.5-2.5 Limited or no protein or DNA contamination shown on gel. |
RIN≥6.5 5.0≥28S/18S≥1.0; limited or no baseline elevation |
Recommended Sample Delivery
Container: 2 ml centrifuge tube (Tin foil is not recommended)
Sample labeling: Group+replicate e.g. A1, A2, A3; B1, B2, B3.
Shipment:
Dry-ice: Samples need to be packed in bags and buried in dry-ice.
RNAstable tubes: RNA samples can be dried in RNA stabilization tube(e.g. RNAstable®) and shipped in room temperature.
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
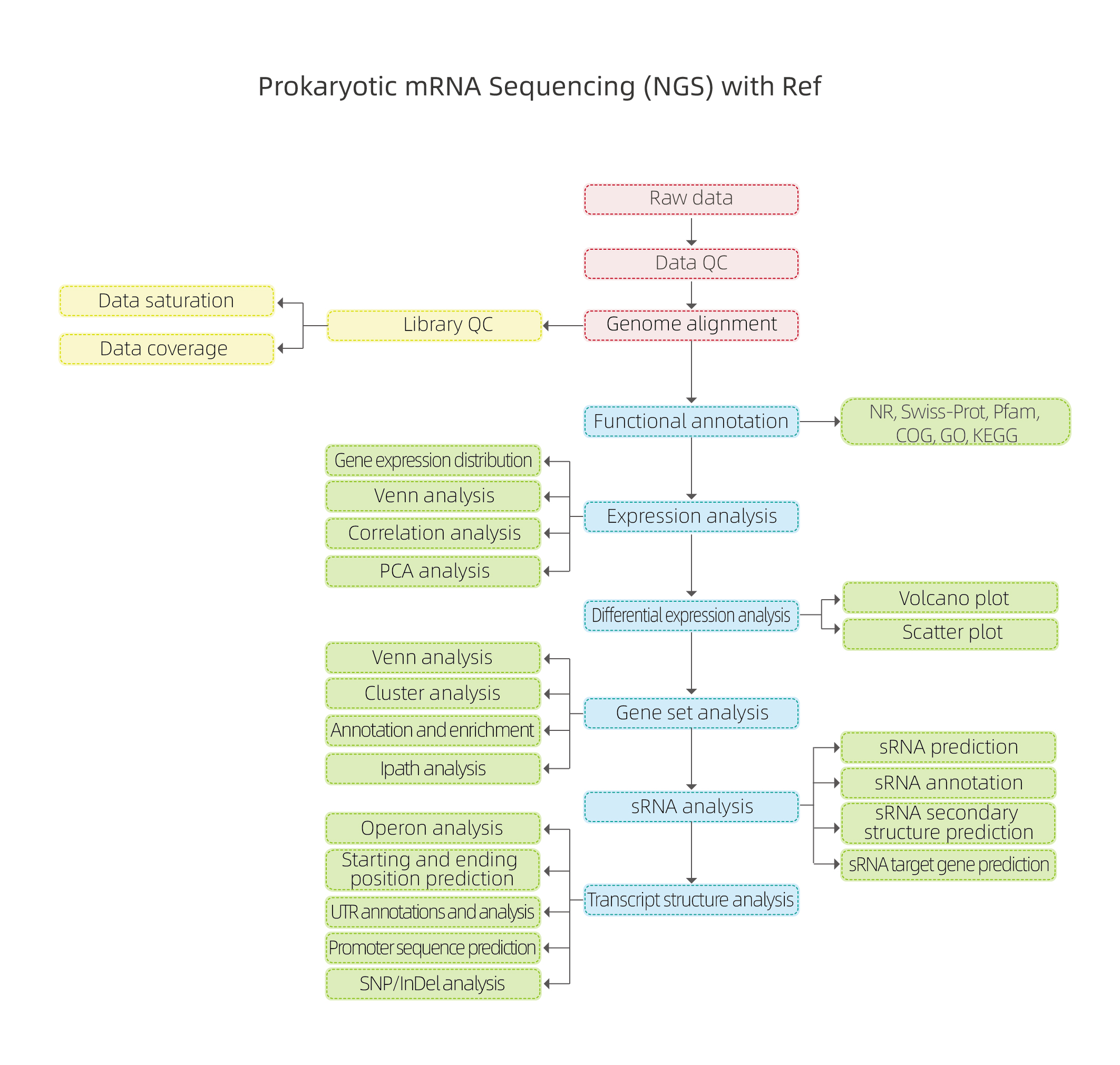
Bioinformatic Analysis Workflow
Includes the following analysis:
● Raw data quality control
● Alignment to the reference genome
● Library quality assessment: RNA fragmentation randomness, insert size and sequencing saturation
● Functional annotation of predicted coding genes
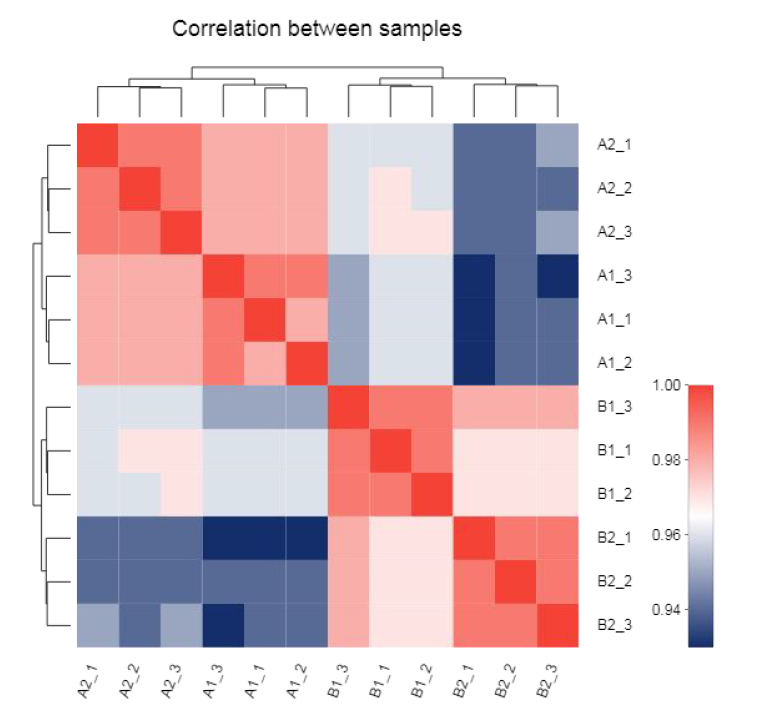
● Expression analysis: correlation and Principal Component Analysis (PCA)
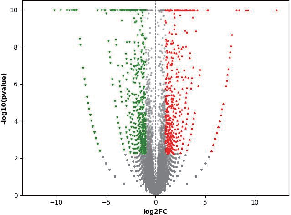
● Differential Gene Expression (DEGs)
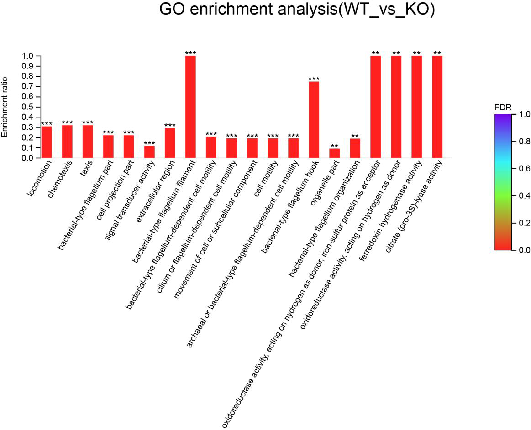
● Functional annotation and enrichment of DEGs
● sRNA analysis: prediction, annotation, target, and secondary structure prediction
● Transcript structure analysis: operons, starting and ending positions, Untranslated region (UTS), promoter, and SNP/InDel analysis
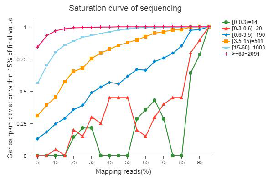
Sequencing saturation
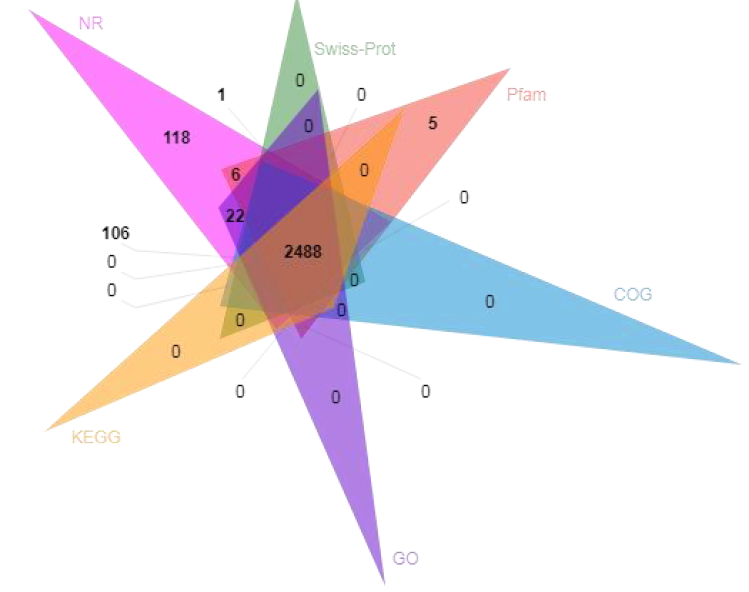
Functional annotation of coding genes
Correlation between samples
Differential Expressed Genes (DEGs) analysis
Functional enrichment analysis
sRNA annotation
Explore the advancements facilitated by BMKGene’s Nanopore full-length mRNA sequencing services in this featured publication.
Guan, C. P. et al. (2018) ‘Global Transcriptome Changes of Biofilm-Forming Staphylococcus epidermidis Responding to Total Alkaloids of Sophorea alopecuroides’, Polish Journal of Microbiology, 67(2), p. 223. doi: 10.21307/PJM-2018-024.