Bacterial complete genome
Service Features
● With 2 possible options to choose from depending on the desired degree of genome completeness:
● Draft genome option: short-read sequencing with Illumina NovaSeq PE150.
● Complete 0 gap genome:
● Long read sequencing on Nanopore Promethion P48 or PacBio Revio for genome assembly.
● Short read sequencing on Illumina NovaSeq for genome validation and error correction (Nanopore) or just to generate a draft genome.
Service Advantages
● 0 gap genome guaranteed: due to the integration of Illumina sequencing with long read sequencing (Nanopore or PacBio).
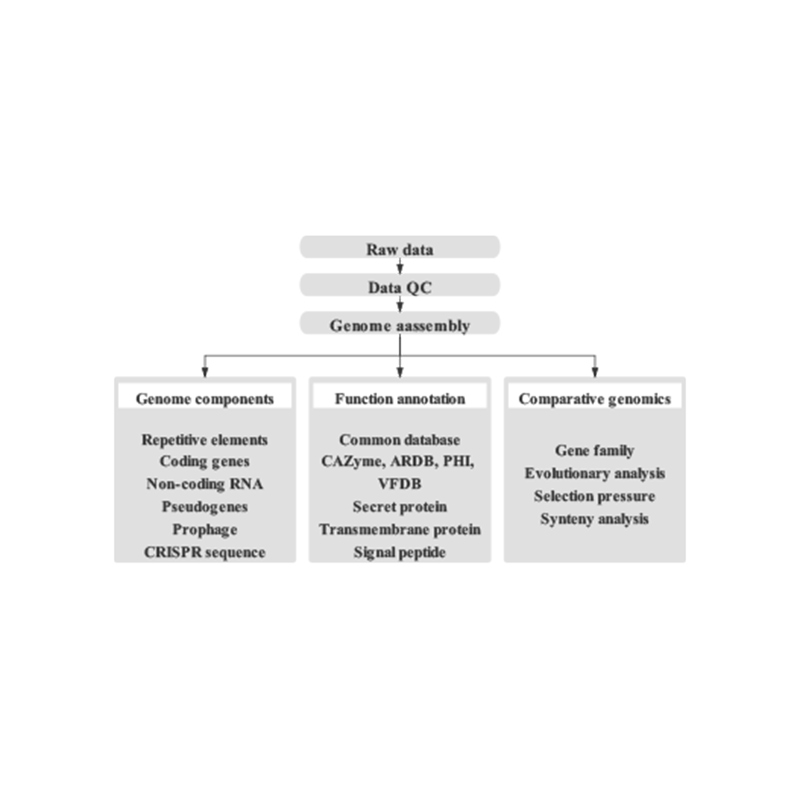
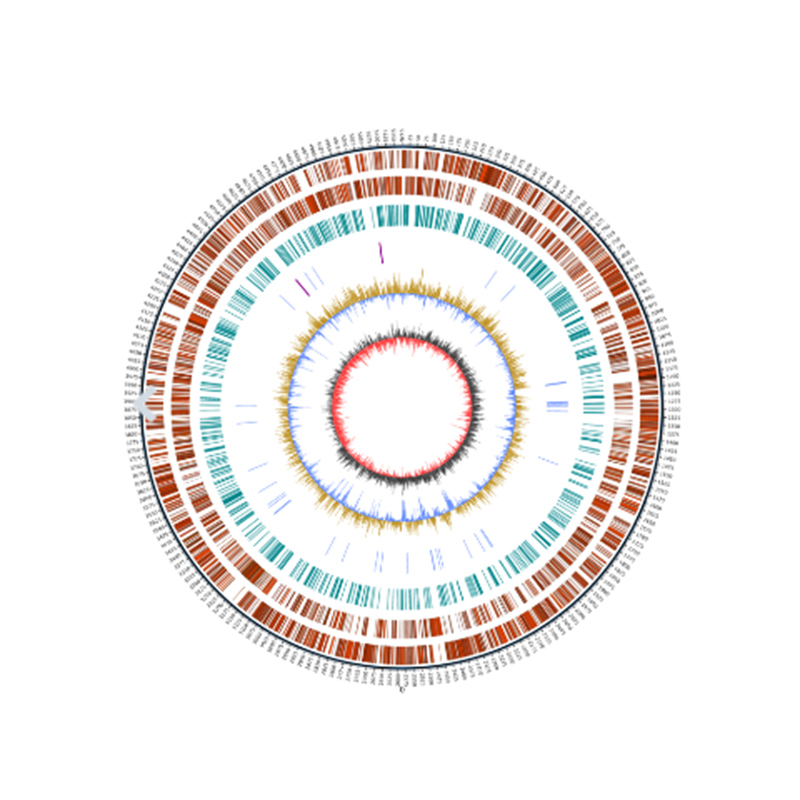
● Complete bioinformatics workflow: including not only genome assembly but also prediction of multiple genomic elements, functional gene annotation, and genome visualisations such as Circos plot.
● Extensive Expertise: with over 10,000 microbial genomes assembled, BMKGENE brings over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
● Multiple sequencing strategies available: for different research goals and requirements of genome completeness
Service Specifications
|
Service |
Sequencing Strategy |
|
Draft Genome |
Illumina PE150 100x |
|
0 Gap Genome |
Nanopore 100x + Illumina PE150 50x Or Pacbio HiFi 30x + Illumina PE150 50x (optional) |
Sample Requirements:
|
Concentration (ng/µL) |
Total amount (µg) |
Volume (µL) |
OD260/280 |
OD260/230 |
|
|
PacBio |
≥20 |
≥1.2 |
≥20 |
1.7-2.2 |
≥1 |
|
Nanopore |
≥40 |
≥2 |
≥20 |
1.7-2.2 |
≥1 |
|
Illumina |
≥1 |
≥0.06 |
≥20 |
1.7-2.2 |
- |
· Bacteria: ≥3.5x1010 cells
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
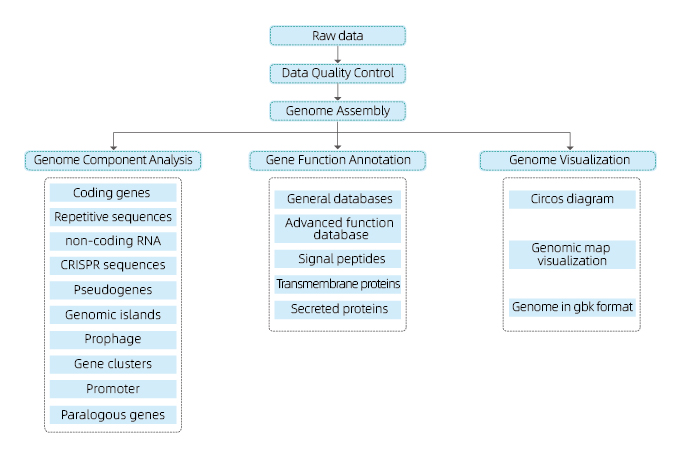
Includes the following analysis:
● Sequencing data quality control
● Genome assembly
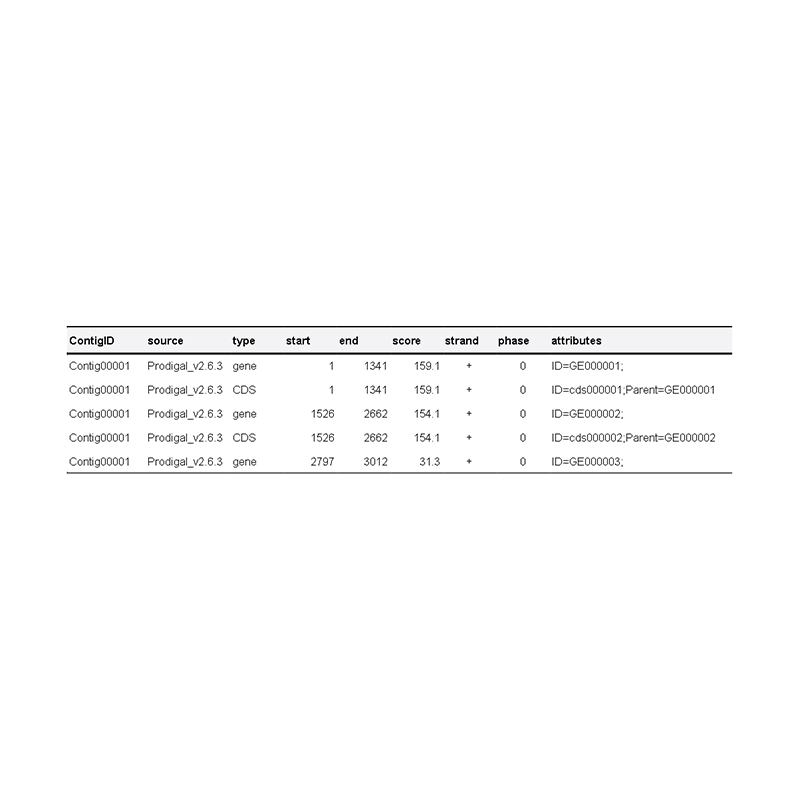
● Genome component analysis: prediction of CDS and multiple genomic elements
● Functional annotation with multiple general databases (GO, KEGG, etc) and advanced databases (CARD, VFDB, etc)
● Genome visualization
Delivery of genome in fasta format and genome annotation file (gff).
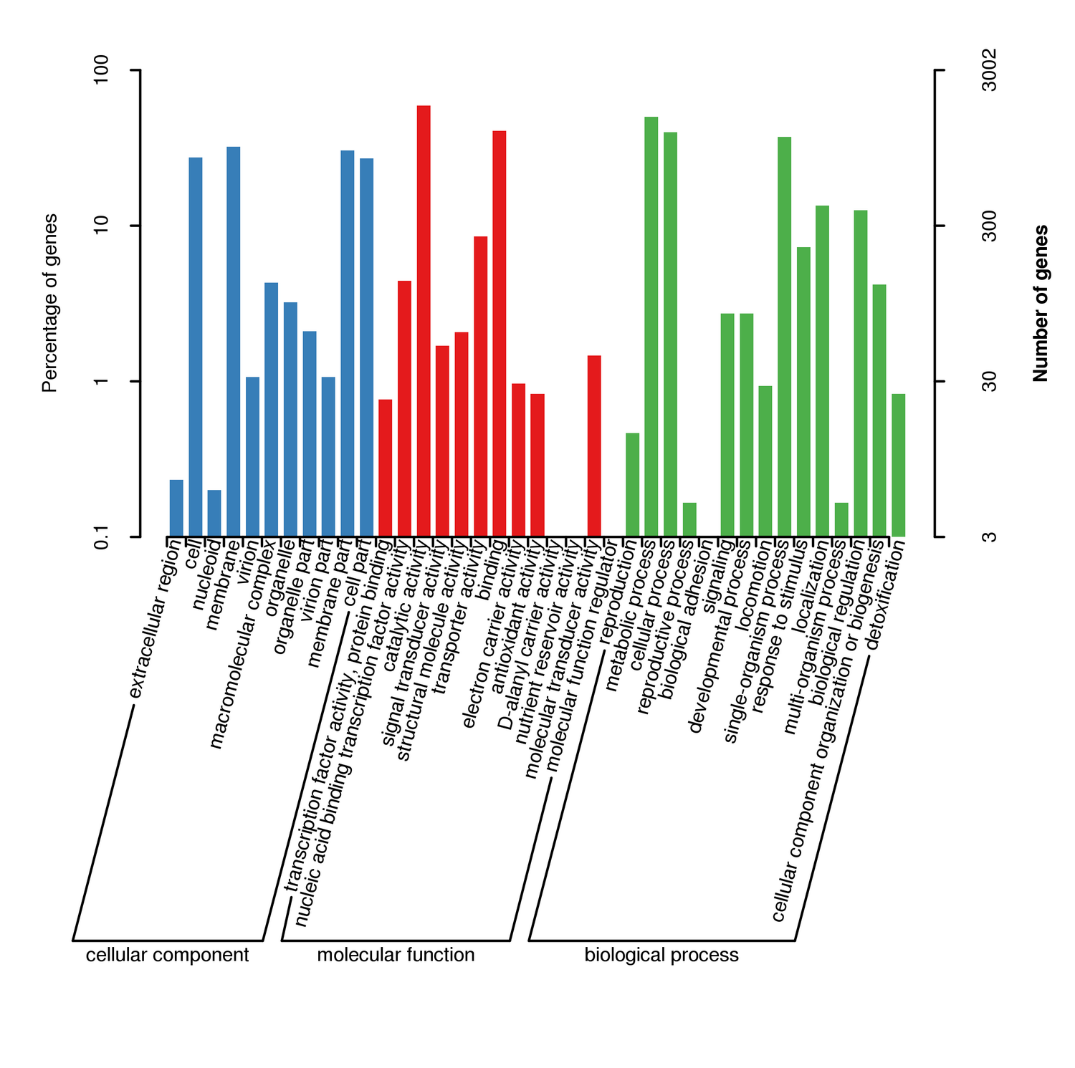
Gene annotation – GO
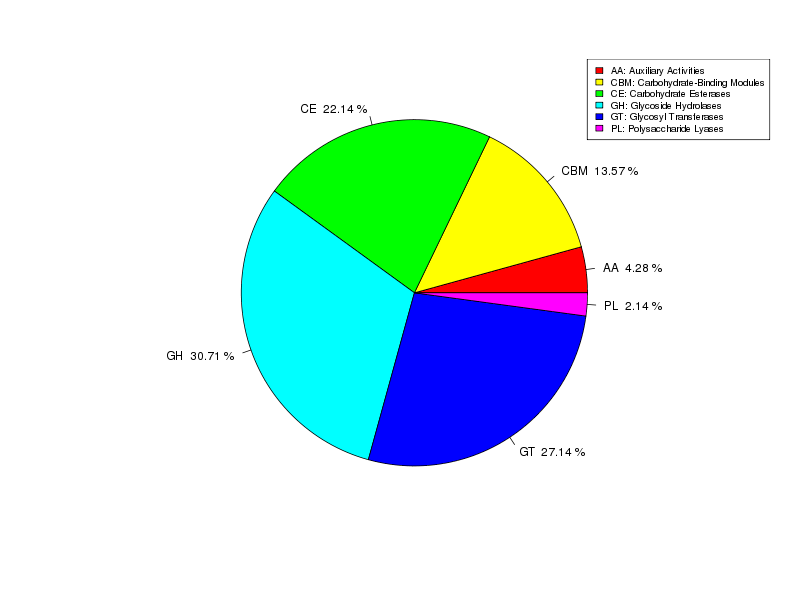
 Gene annotation – CAZY carbohydrates
Gene annotation – CAZY carbohydrates
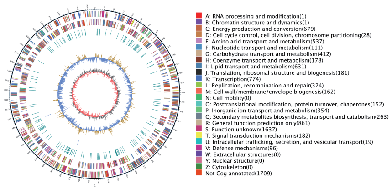
Genome visualization – Circos plot
Explore the advancements facilitated by BMKGene’s bacterial genome assembly services through a curated collection of publications.
Dai, W. et al. (2023) ‘Discovery of Bacteroides uniformis F18-22 as a Safe and Novel Probiotic Bacterium for the Treatment of Ulcerative Colitis from the Healthy Human Colon’, International Journal of Molecular Sciences, 24(19), p. 14669. doi: 10.3390/IJMS241914669/S1.
Kang, Q. et al. (2021) ‘Multidrug-resistant Proteus mirabilis isolates carrying blaOXA-1 and blaNDM-1 from wildlife in China: increasing public health risk’, Integrative Zoology, 16(6), pp. 798–809. doi: 10.1111/1749-4877.12510.
Wang, T. T. et al. (2017) ‘Complete genome sequence of endophyte Bacillus flexus KLBMP 4941 reveals its plant growth promotion mechanism and genetic basis for salt tolerance’, Journal of Biotechnology, 260, pp. 38–41. doi: 10.1016/J.JBIOTEC.2017.09.001.
Wang, X. et al. (2021) ‘Multiple-Replicon Resistance Plasmids of Klebsiella Mediate Extensive Dissemination of Antimicrobial Genes’, Frontiers in Microbiology, 12, p. 754931. doi: 10.3389/FMICB.2021.754931/BIBTEX.