circRNA sequencing-Illumina
Features
● rRNA depletion followed by directional library preparation, enabling strand-specific sequencing data.
● Bioinformatic workflow enables circRNA prediction and expression quantification
Service Advantages
● More comprehensive RNA libraries: we use rRNA depletion instead of linear RNA depletion in our pre-library preparation, ensuring that the sequencing data includes not only circRNA but also mRNA and lncRNA, enabling joint analysis of these datasets
● Optional analysis of competitive endogenous RNA (ceRNA) networks: providing deeper insights into cellular regulatory mechanisms
● Extensive Expertise: with a track record of processing over 20,000 samples at BMK, spanning diverse sample types and lncRNA projects, our team brings a wealth of experience to every project.
● Rigorous Quality Control: we implement core control points across all stages, from sample and library preparation to sequencing and bioinformatics. This meticulous monitoring ensures the delivery of consistently high-quality results.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Sample Requirements and Delivery
|
Library |
Platform |
Recommended data |
Data QC |
|
Poly A enriched |
Illumina PE150 |
16-20 Gb |
Q30≥85% |
Sample Requirements:
Nucleotides:
|
Conc.(ng/μl) |
Amount (μg) |
Purity |
Integrity |
|
≥ 100 |
≥ 0.5 |
OD260/280=1.7-2.5 OD260/230=0.5-2.5 Limited or no protein or DNA contamination shown on gel. |
For plants: RIN≥6.5; For animals: RIN≥7.0; 5.0≥28S/18S≥1.0; limited or no baseline elevation |
● Plants:
Root, Stem or Petal: 450 mg
Leaf or Seed: 300 mg
Fruit: 1.2 g
● Animal:
Heart or Intestine: 450 mg
Viscera or Brain: 240 mg
Muscle: 600 mg
Bones, Hair or Skin: 1.5g
● Arthropods:
Insects: 9g
Crustacea: 450 mg
● Whole blood: 2 tubes
● Cells: 106 cells
● Serum and Plasma: 6 mL
Recommended Sample Delivery
Container: 2 ml centrifuge tube (Tin foil is not recommended)
Sample labeling: Group+replicate e.g. A1, A2, A3; B1, B2, B3.
Shipment:
1. Dry-ice: Samples need to be packed in bags and buried in dry-ice.
2. RNAstable tubes: RNA samples can be dried in RNA stabilization tube(e.g., RNAstable®) and shipped at room temperature.
Service Work Flow

Experiment design

Sample delivery

RNA extraction

Library construction

Sequencing

Data analysis

After-sale services
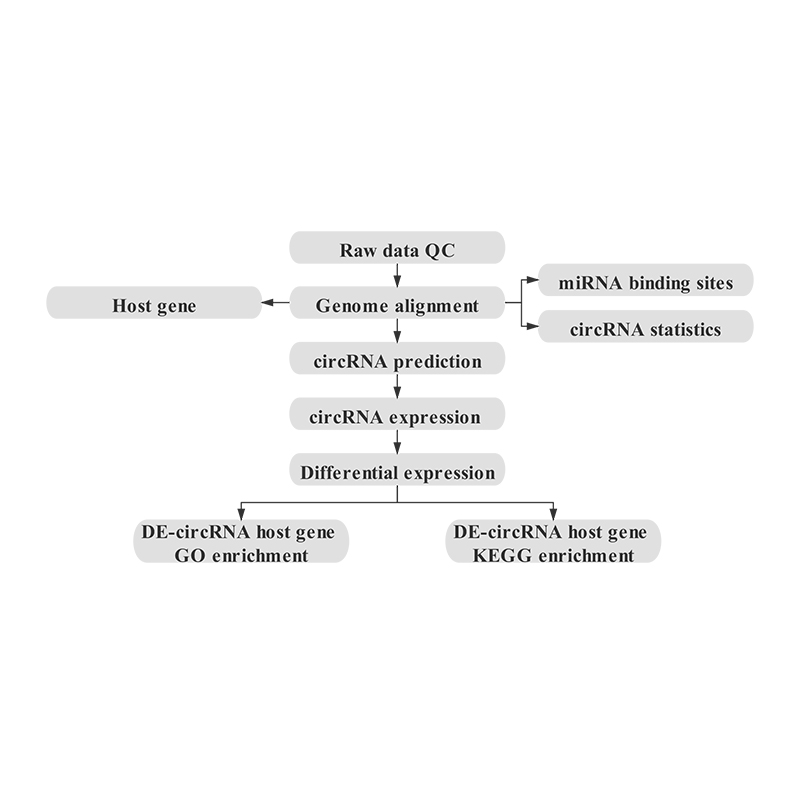
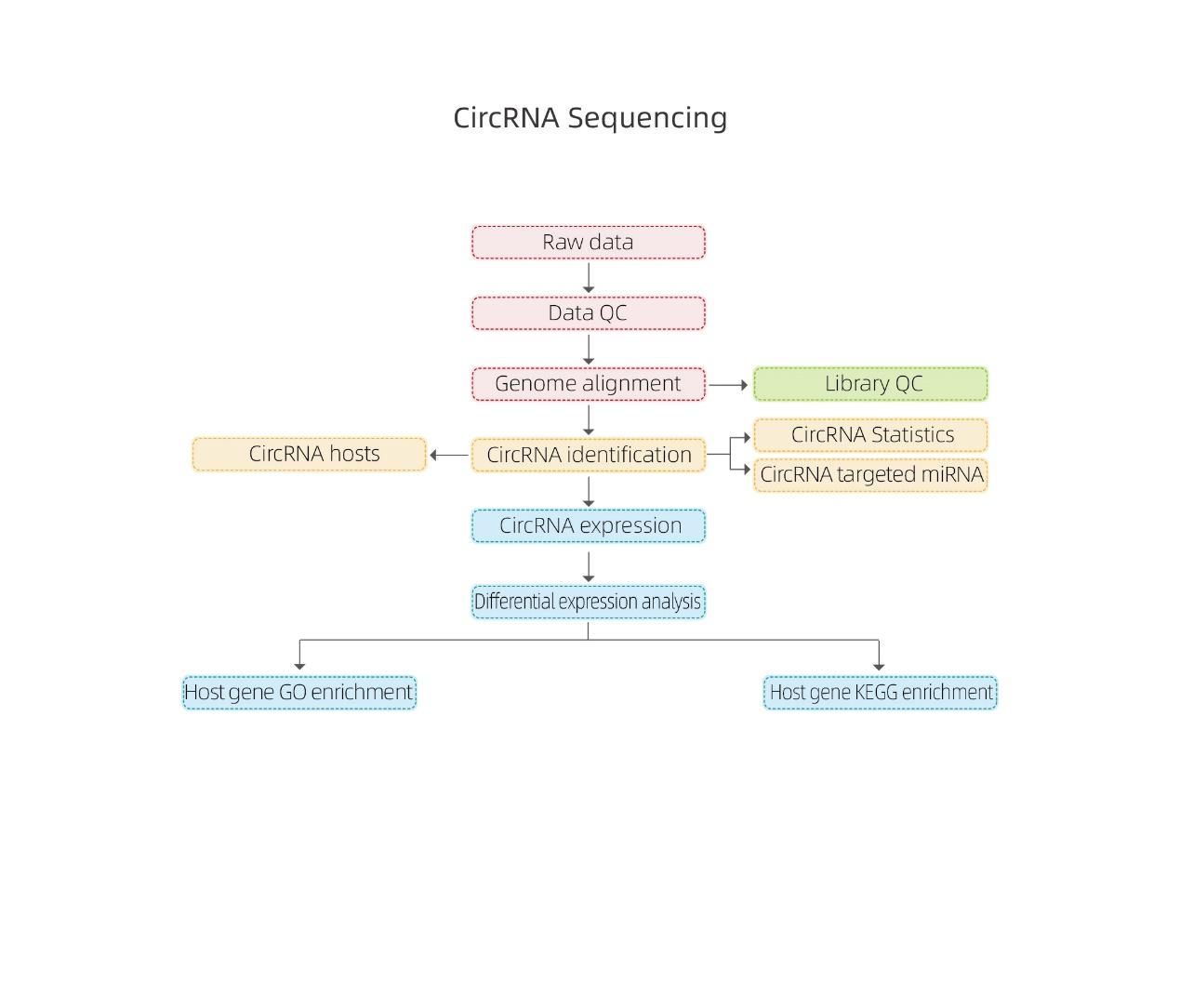
Bioinformatics
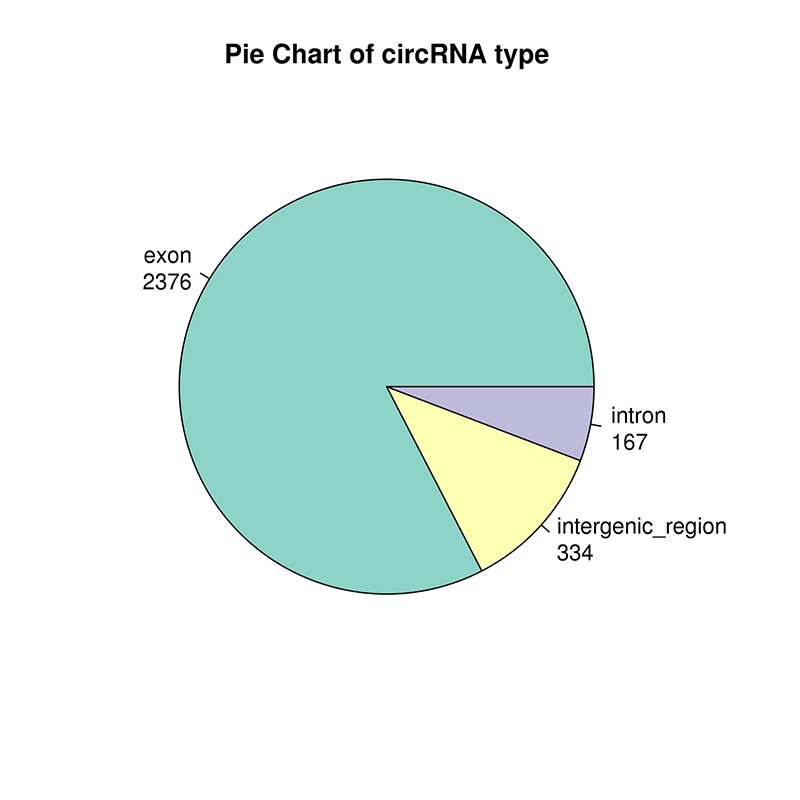
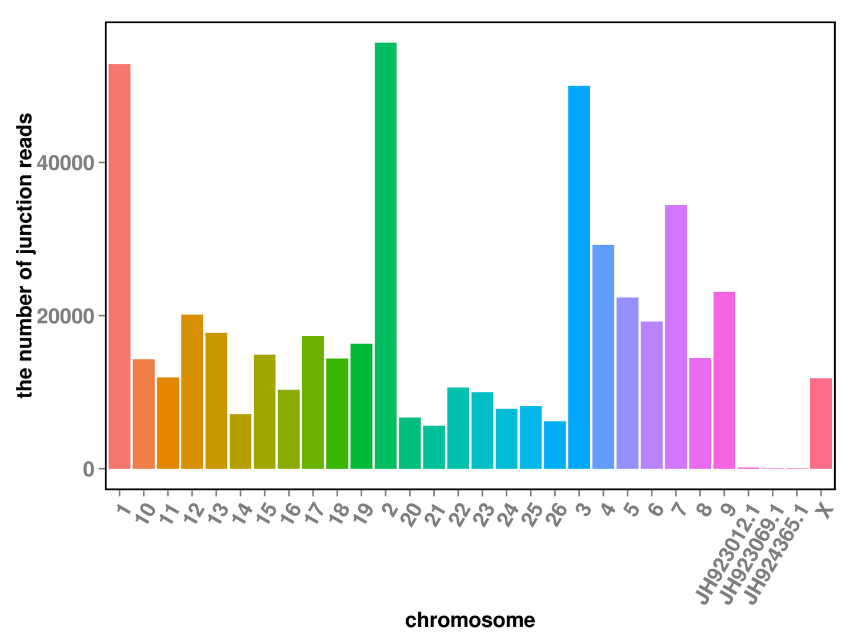
circRNA prediction: chromosomal distribution
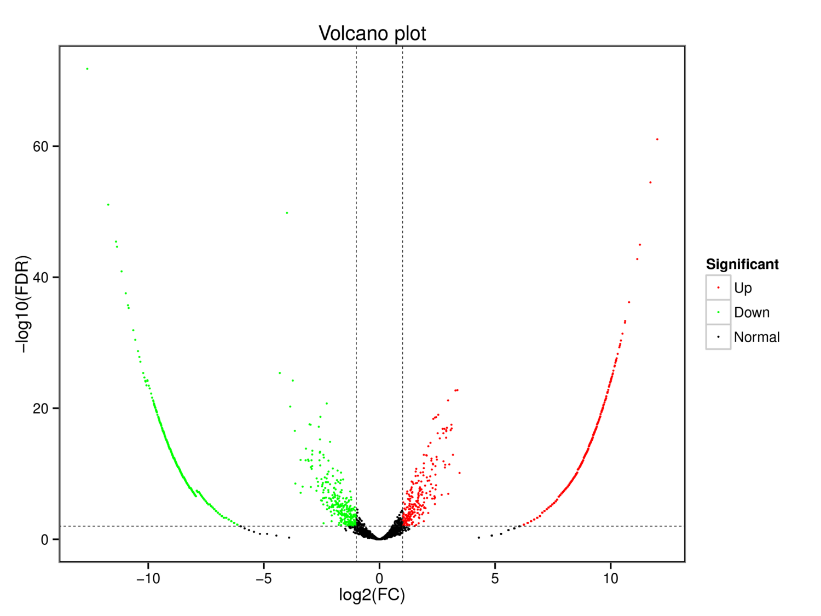
Differentially Expressed circRNAs – Volcano plot
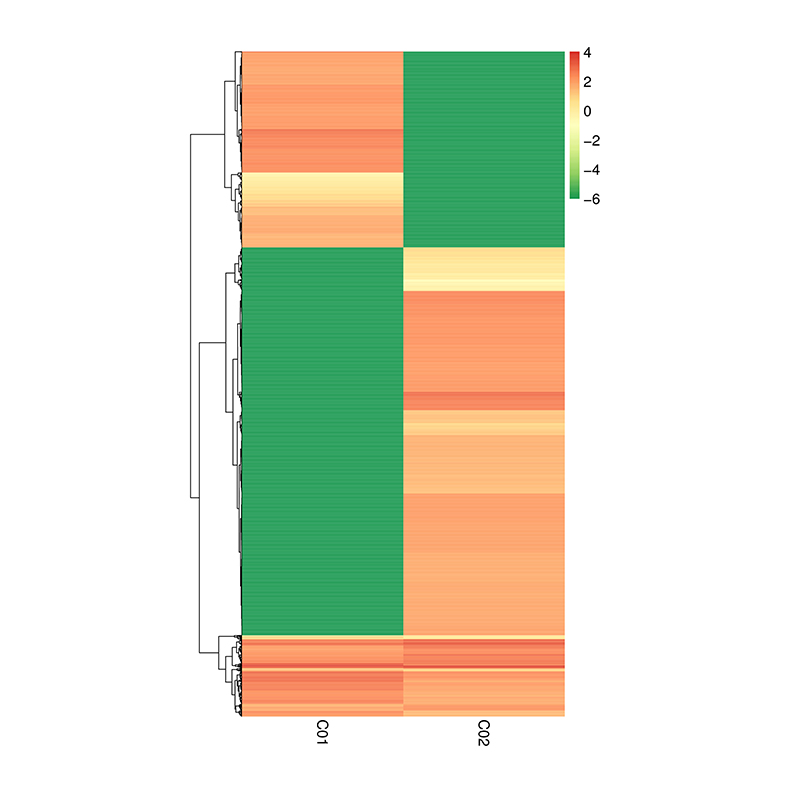
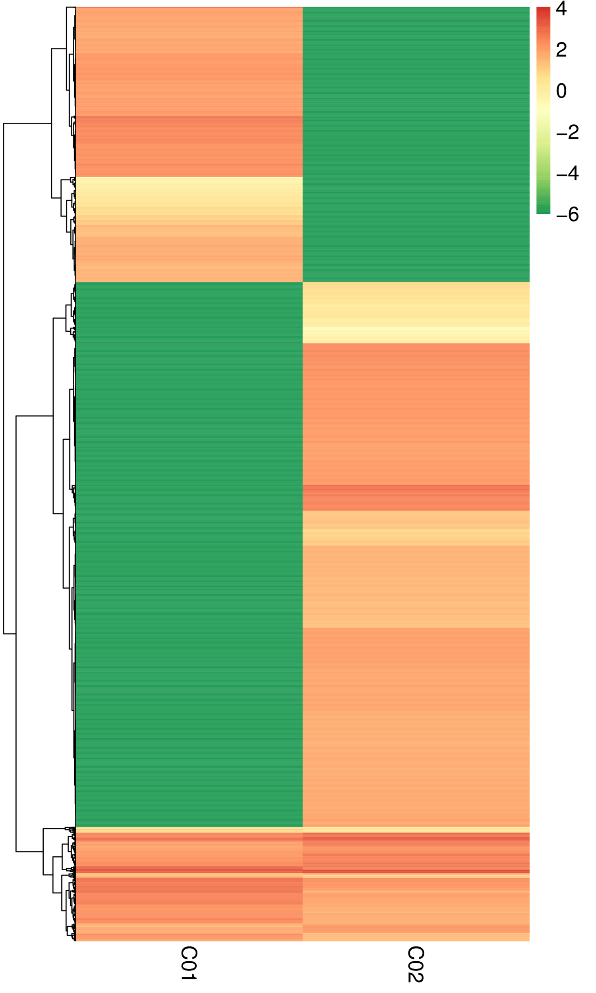
Differentially Expressed circRNAs – hierarchical clustering
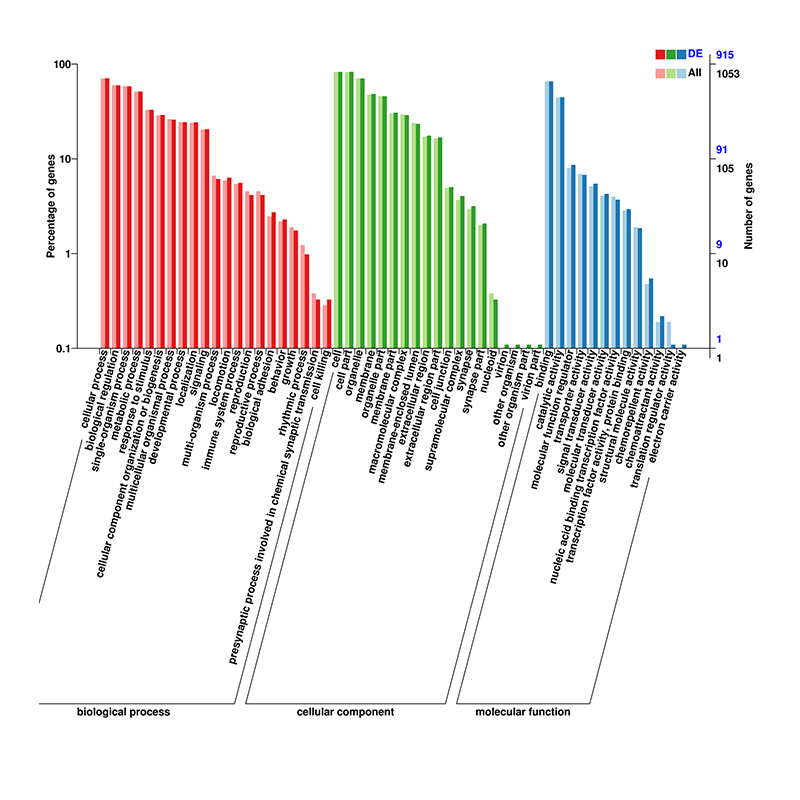
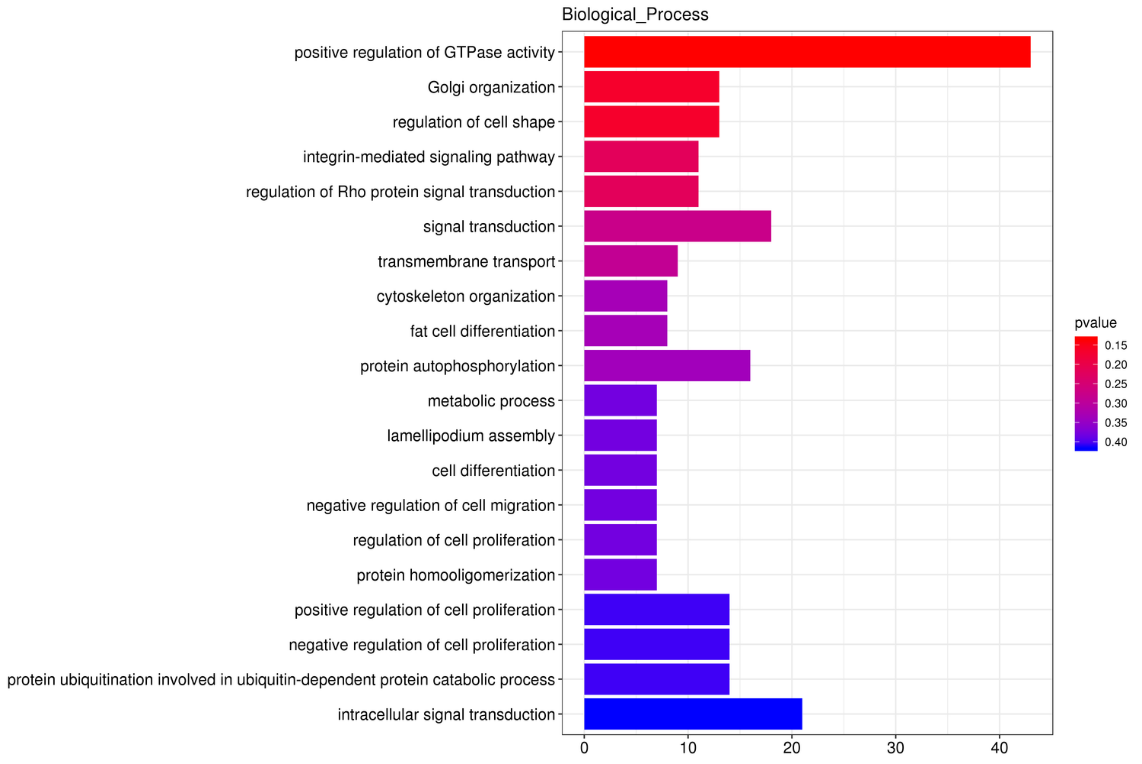
Functional enrichment of circRNA’s host genes
Explore the research advancements facilitated by BMKGene’ circRNA sequencing services through a curated collection of publications.
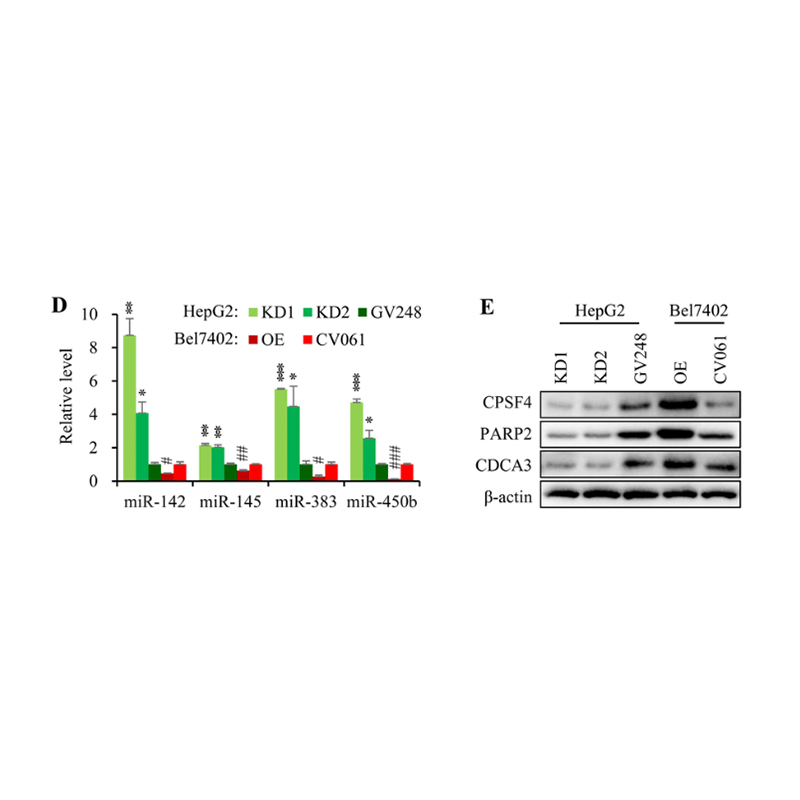
Wang, X. et al. (2021) ‘CPSF4 regulates circRNA formation and microRNA mediated gene silencing in hepatocellular carcinoma’, Oncogene 2021 40:25, 40(25), pp. 4338–4351. doi: 10.1038/s41388-021-01867-6.
Xia, K. et al. (2023) ‘X oo-responsive transcriptome reveals the role of the circular RNA133 in disease resistance by regulating expression of OsARAB in rice’, Phytopathology Research, 5(1), pp. 1–14. doi: 10.1186/S42483-023-00188-8/FIGURES/6.
Y, H. et al. (2023) ‘CPSF3 modulates the balance of circular and linear transcripts in hepatocellular carcinoma’. doi: 10.21203/RS.3.RS-2418311/V1.
Zhang, Y. et al. (2023) ‘Comprehensive evaluation of circRNAs in cirrhotic cardiomyopathy before and after liver transplantation’, International Immunopharmacology, 114, p. 109495. doi: 10.1016/J.INTIMP.2022.109495.