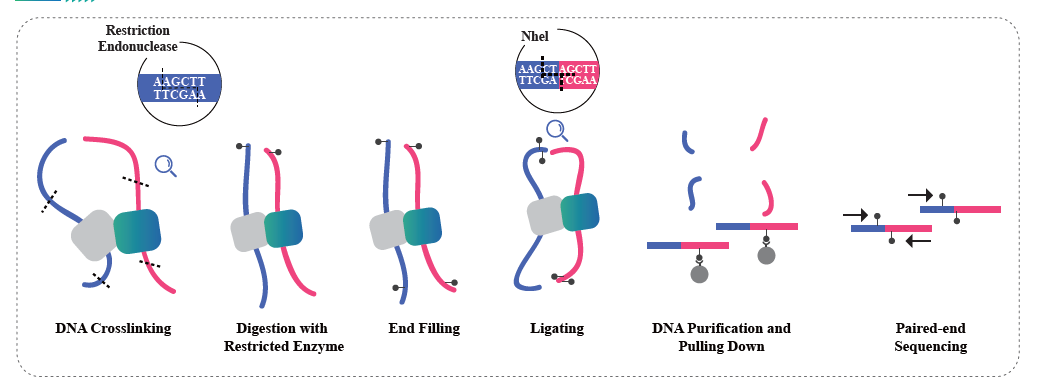
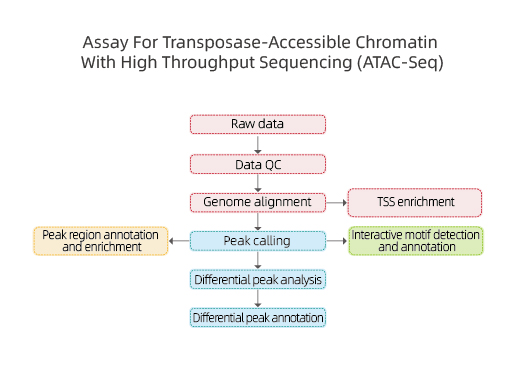
Assay for Transposase-Accessible Chromatin with High Throughput Sequencing (ATAC-seq)
Service Features
● Service requires tissue samples, instead of extracted nucleic acids, to conserve the DNA-protein interactions.
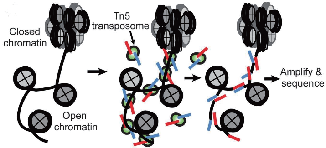
● ATAC method includes tissue dissociation, followed by nuclei isolation, Tn5 treatment and purification, PCR amplification, size selection and sequencing.
● Sequencing in Illumina NovaSeq
Service Advantages
● High sensitivity: low starting cell quantity is sufficient for library preparation and sequencing.
● Highly informative technique: simultaneously reveals the genomic locations of open chromatin and active regulatory elements such as transcription factor binding sites.
● Good experimental reproducibility: technical replicates show excellent repeatability.
● Extensive Expertise: with hundreds of ATAC sequencing projects successfully completed, BMKGENE brings over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
● Possibility to join with transcriptomics analysis: allowing for the integrated analysis of ATAC seq with other omics data such as RNA-seq.
· Comprehensive bioinformatic analysis: enabling not only the identification of the open regions of the chromatin and their corresponding functionality (promoters, UTs, exons, introns) but also the analysis of differences between chromatin open regions among samples.
Service Specifications
|
Library |
Sequencing Strategy |
Recommended data output |
Quality control |
|
ATAC-seq |
Illumina PE150 |
> 20M reads Depending on genome size (Human: 50 M reads) |
Q30≥85% |
Sample Requirements
Samples type: Tissues, live or frozen cells, blood
● Cell number: ≥ 106 cells
● Tissue weight: ≥ 200mg fresh tissue
● Blood: ≥ 4 mL
Service Work Flow

Experiment design

Sample delivery

DNA extraction

Library construction

Sequencing

Data analysis

After-sale services
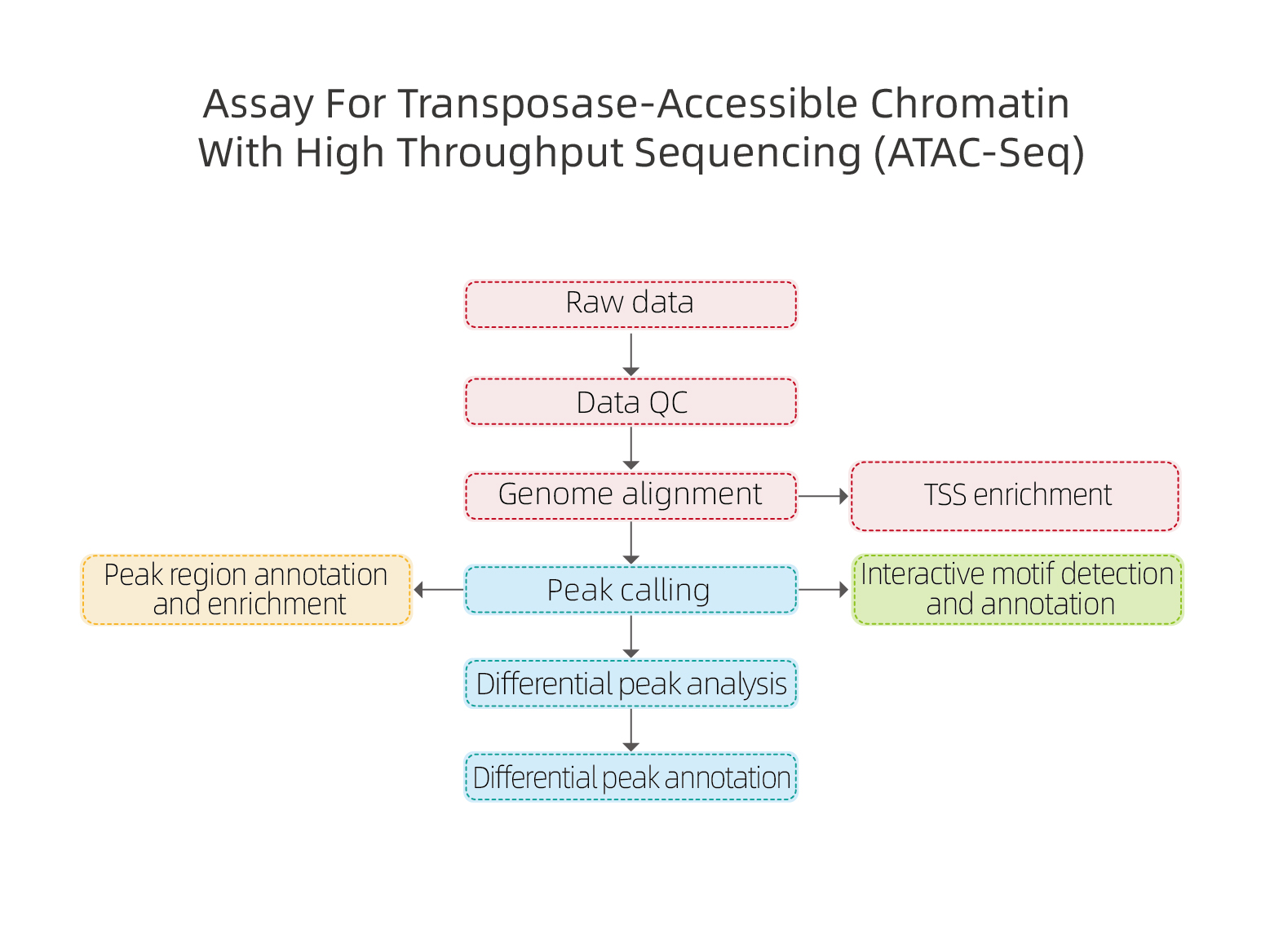
Includes the following analysis:
● Raw data quality control;
● Peak calling based on mapping to reference genome;
● Gene annotation on peak location;
● Motif analysis: identification of transcription factor binding sites (TFBS);
● Differential Peak Analysis and annotation.
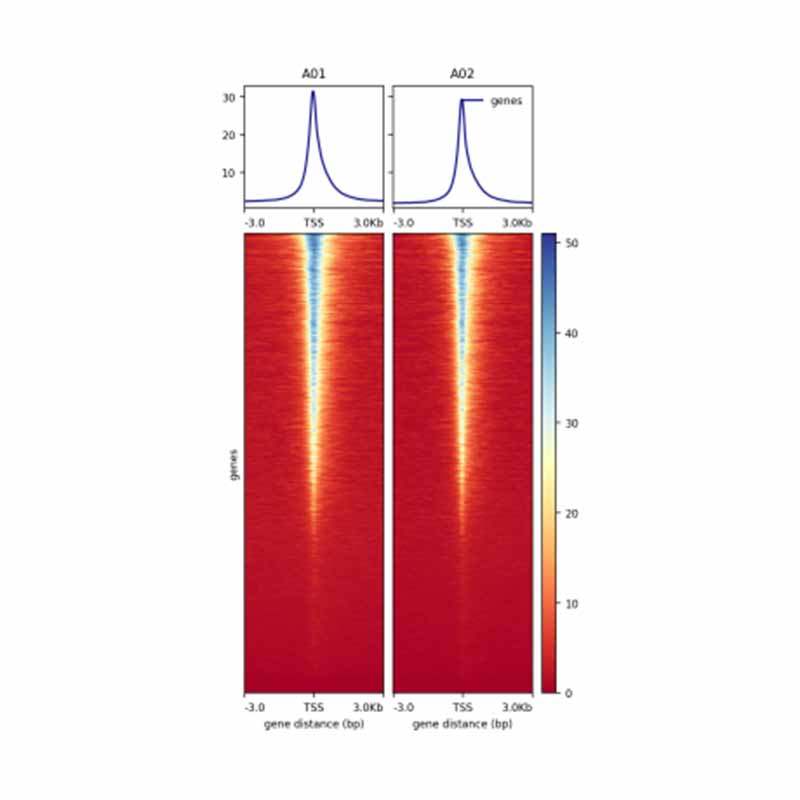
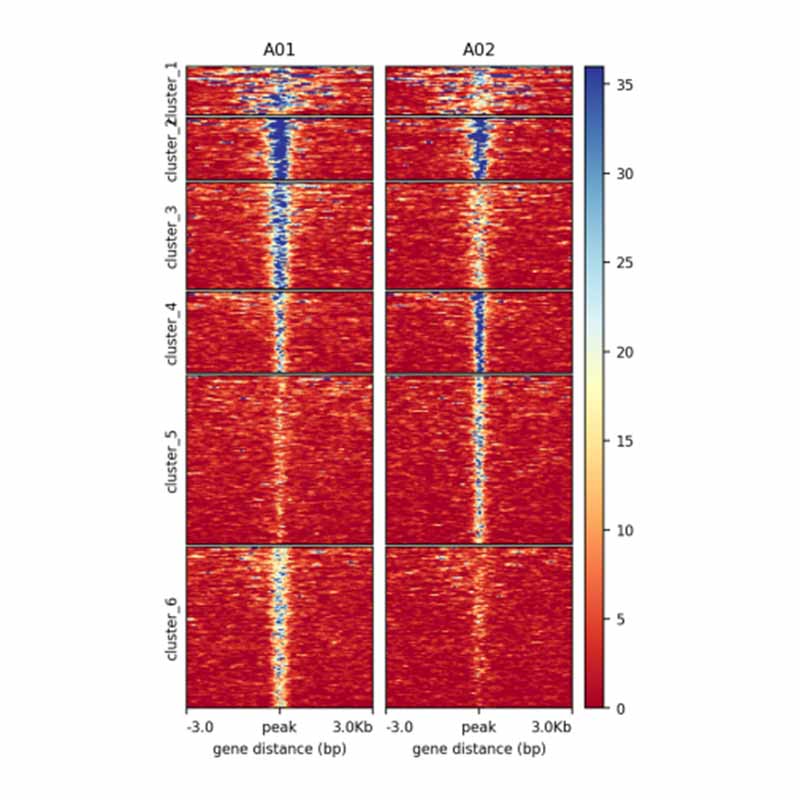
1.Heatmap on ATAC reads distribution at TSS and adjacent regions (±3 kb)
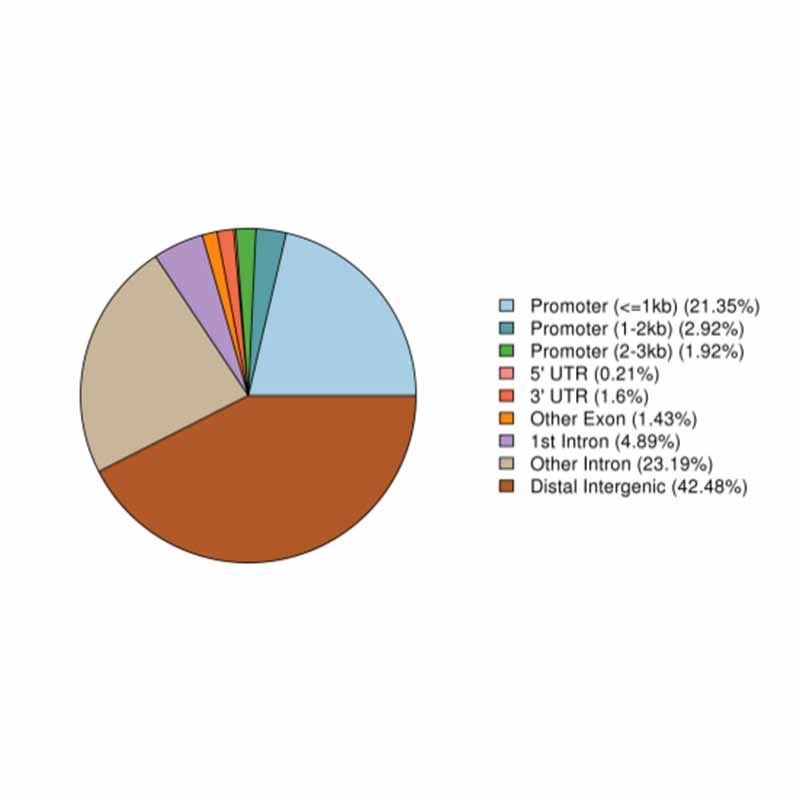
1.jpg) 2.Distribution of open chromatin region in different genome components
2.Distribution of open chromatin region in different genome components
3.Differential peak calling between groups
Explore the research advancements facilitated by BMKGene’s ATAC sequencing services through a curated collection of publications.
Fu, A. et al. (2023) ‘Telomere-to-telomere genome assembly of bitter melon (Momordica charantia L. var. abbreviata Ser.) reveals fruit development, composition and ripening genetic characteristics’, Horticulture Research, 10(1). doi: 10.1093/HR/UHAC228.
Gong, B. et al. (2023) ‘Epigenetic and transcriptional activation of the secretory kinase FAM20C as an oncogene in glioma’, Journal of Genetics and Genomics, 50(6), pp. 422–433. doi: 10.1016/J.JGG.2023.01.008.
He, Y. et al. (2023) ‘Butyrate reverses ferroptosis resistance in colorectal cancer by inducing c-Fos-dependent xCT suppression’, Redox Biology, 65, p. 102822. doi: 10.1016/J.REDOX.2023.102822.

.jpg)
.jpg)