Evolutionary Genetics
Service Advantages
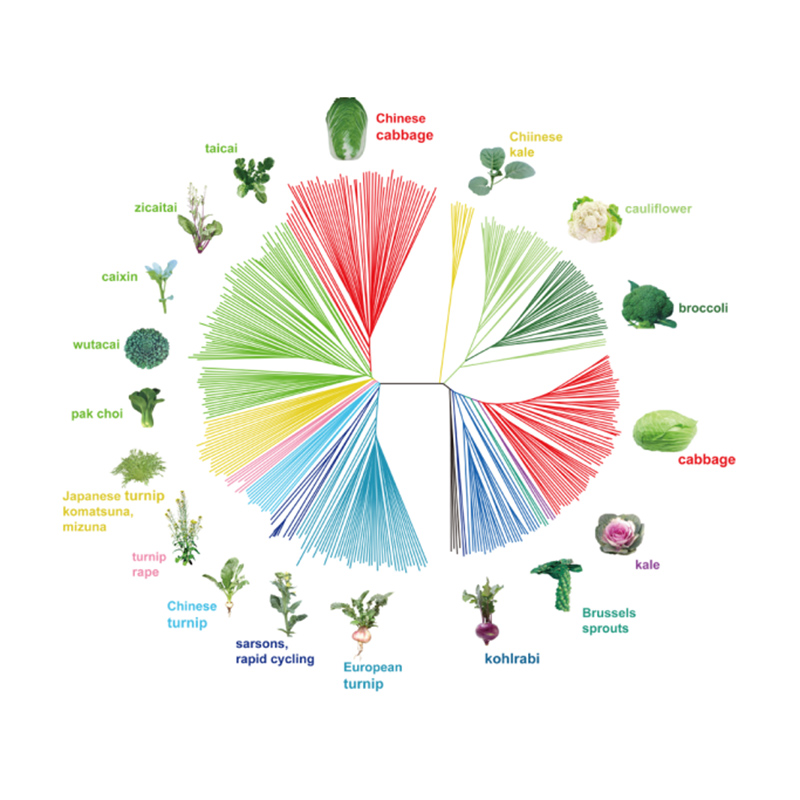
Takagi et al., The plant journal, 2013
● Comprehensive bioinformatic analysis: enabling the estimation of genetic diversity, which reflects the evolutionary potential of species, and revealing reliable phylogenetic relation between species with minimized influence of convergent evolution and parallel evolution
● Optional customized analysis: such as the estimation of divergence time and speed based on variations at nucleotide and amino acids level.
● Extensive Expertise and publication records: BMKGene has accumulated massive experience in population and evolutionary genetics projects for over 12 years, covering hundreds of species, etc. and contributed to over 80 high-level projects published in Nature Communications, Molecular Plants, Plant Biotechnology Journal, etc.
● Highly skilled bioinformatics team and short analysis cycle: with great experience in advanced genomics analysis, BMKGene’s team delivers comprehensive analyses with a swift turnaround time.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Service Specifications and requirements
|
Type of sequencing |
Recommended population scale |
Sequencing strategy |
Nucleotide requirements |
|
Whole Genome Sequencing |
>30 individuals, with >10 individuals from each subgroup
|
10x |
Concentration: ≥1 ng/ µL Total amount≥ 30ng Limited or no degradation or contamination |
|
Specific-Locus Amplified Fragment (SLAF) |
Tag depth: 10x Number of tags: <400 Mb: WGS is recommended <1Gb: 100K tags 1Gb<genome<2Gb: 200K tags >2Gb: 300K tags Max 500k tags |
Concentration ≥ 5 ng/µL Total amount ≥80 ng Nanodrop OD260/280=1.6-2.5 Agarose gel: no or limited degradation or contamination
|
Sample Requirements and Delivery
Sample Requirements:
|
Species |
Tissue |
WGS-NGS |
SLAF |
|
Animal
|
Visceral tissue |
0.5~1g
|
0.5g
|
|
Muscle tissue |
|||
|
Mammalian blood |
1.5mL
|
1.5mL
|
|
|
Poultry/Fish blood |
|||
|
Plant
|
Fresh Leaf |
1~2g |
0.5~1g |
| Petal/Stem | |||
| Root/Seed | |||
|
Cells |
Cultured cell |
|
gDNA |
Concentration |
Amount (ug) |
OD260/OD280 |
|
SLAF |
≥35 |
≥1.6 |
1.6-2.5 |
|
WGS-NGS |
≥1 |
≥0.1 |
- |
Service Work Flow

Experiment design

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
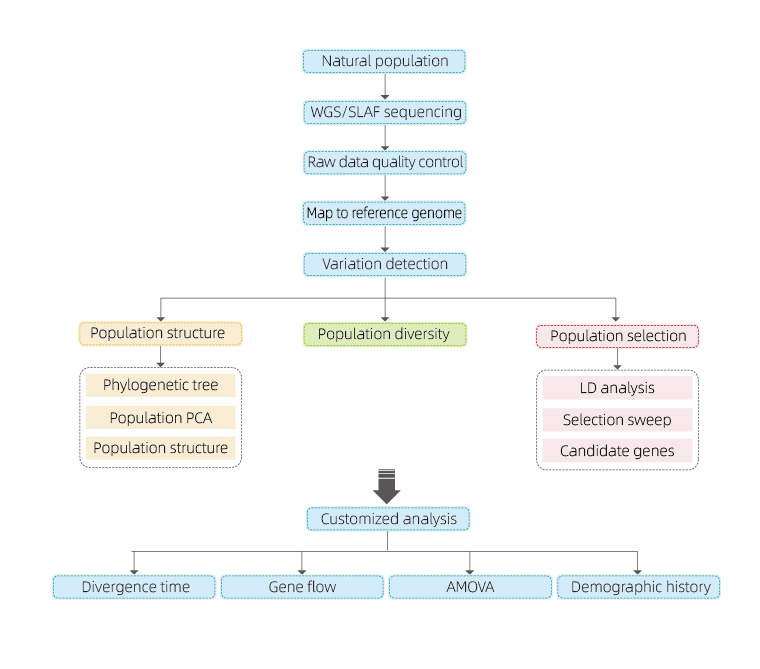
Service includes analysis of population structure (phylogenetic tree, PCA, population stratification chart), population diversity, and population selection (linkage disequilibrium, selective sweep-selection of advantageous sites). The service can also included customized analysis (e.g. divergence time, gene flow).
*Demo results shown here are all from genomes published with BMKGENE
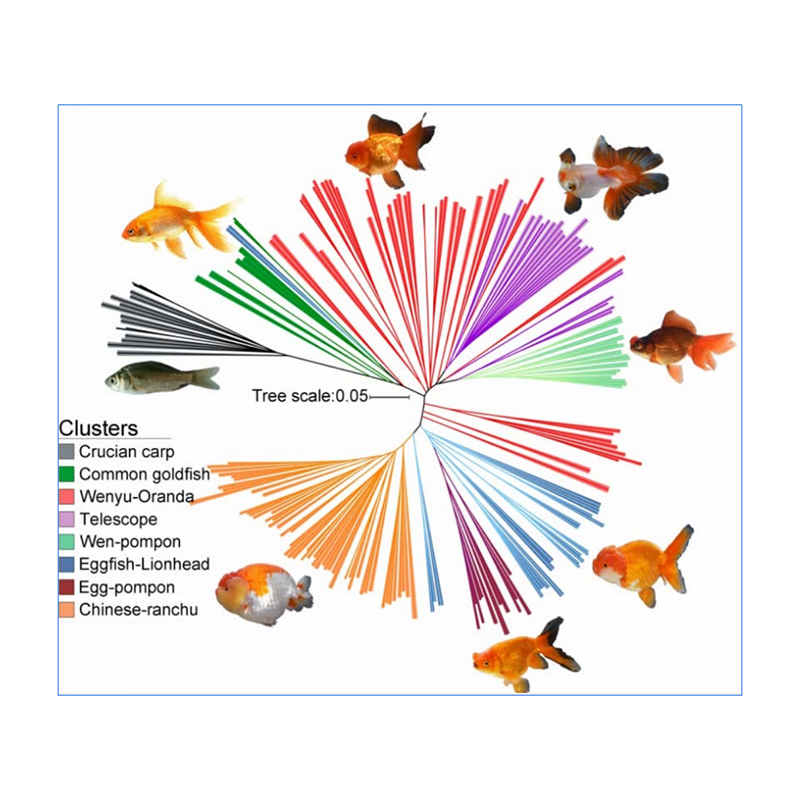
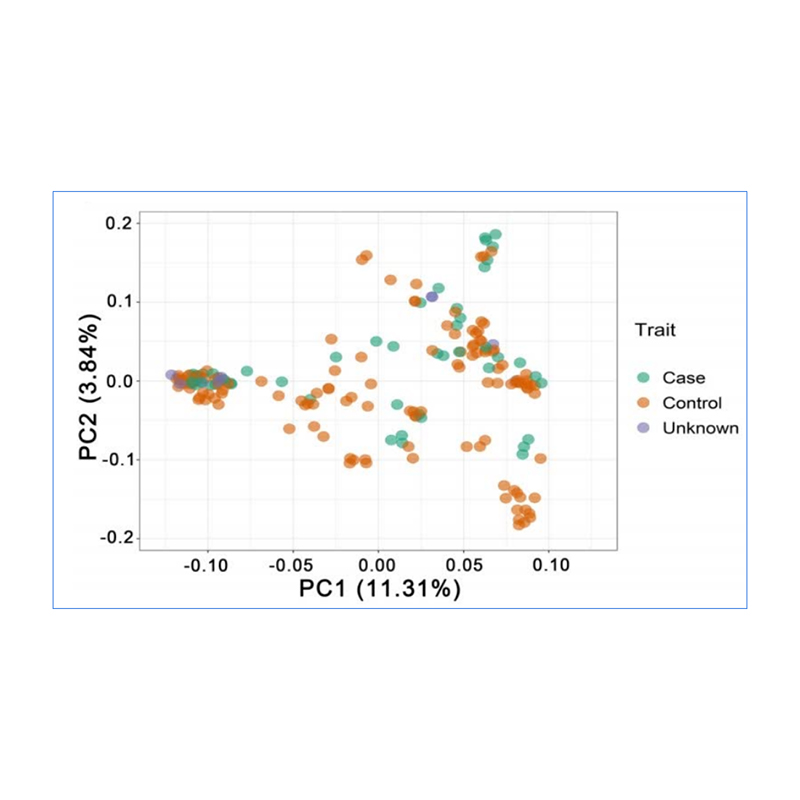
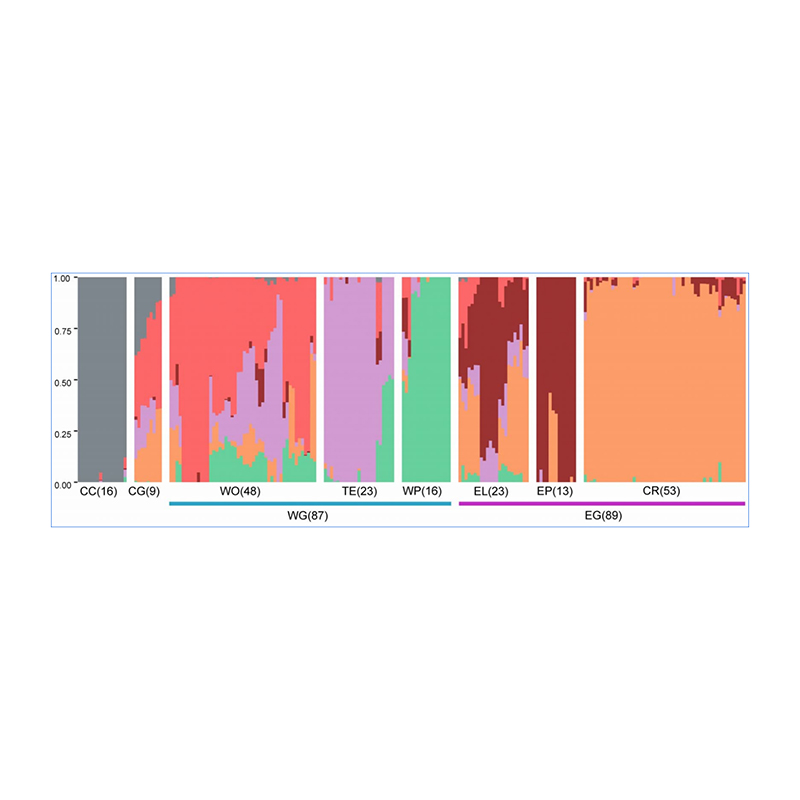
1.Evolution analysis contains construction of phylogenetic tree, population structure and PCA based on genetic variations.
Phylogenetic tree represents taxonomic and evolutionary relationships among species with common ancestor.
PCA aims to visualize closeness between sub-populations.
Population structure shows the presence of genetically distinct sub-population in terms of allele frequencies.
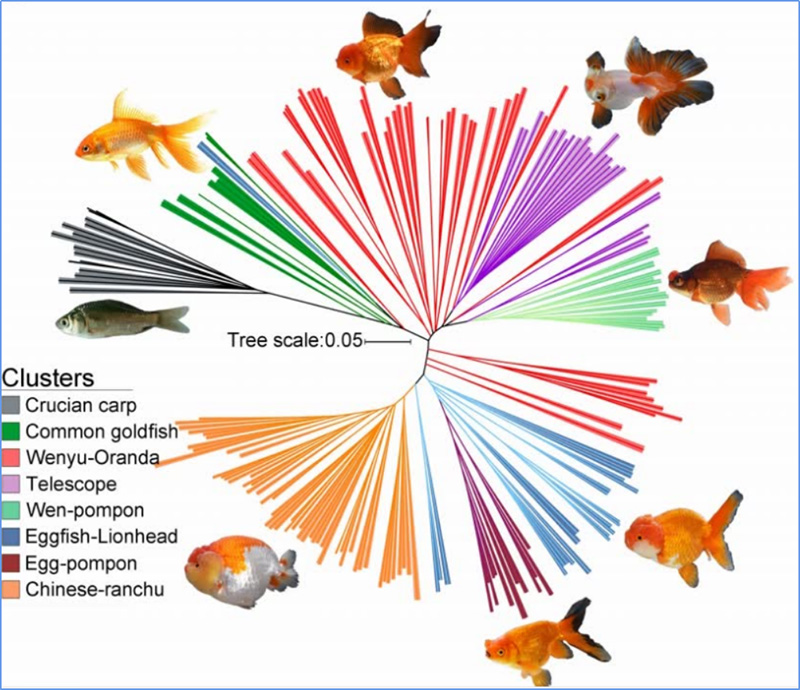
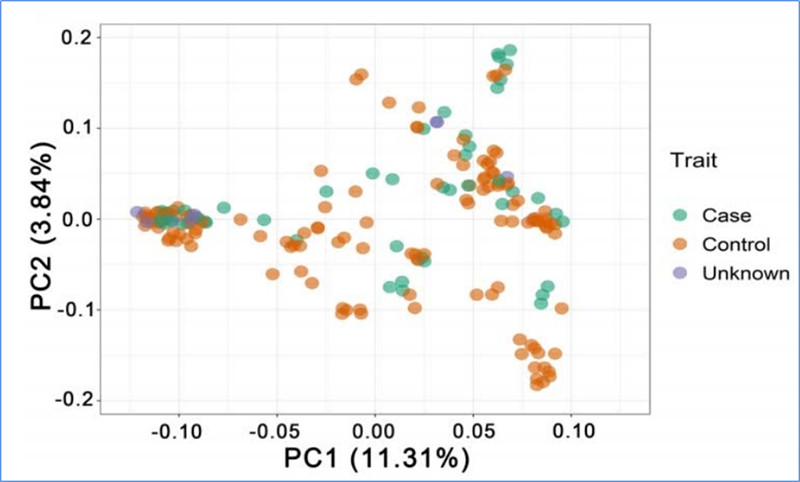
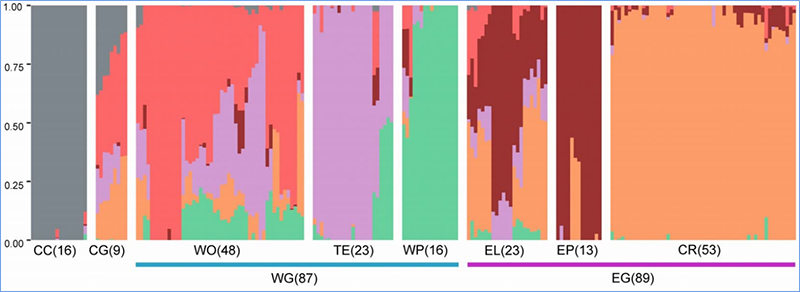
Chen, et. al., PNAS, 2020
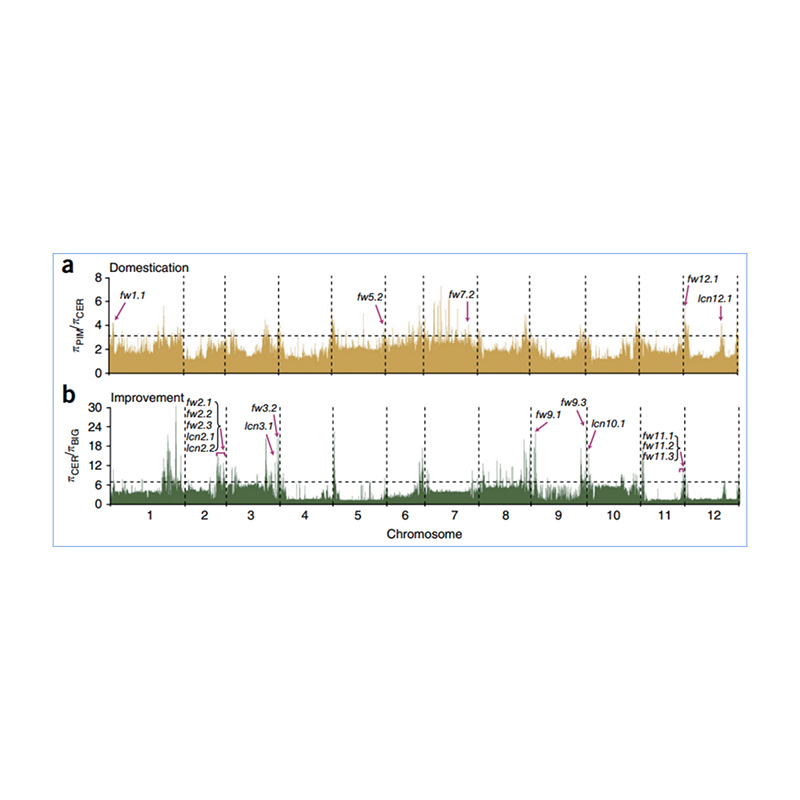
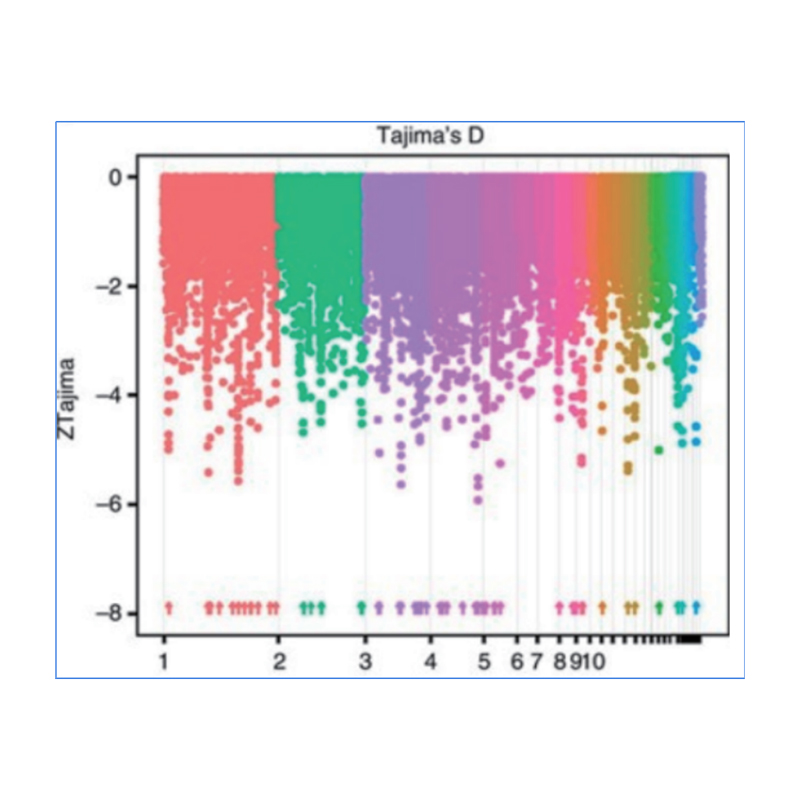
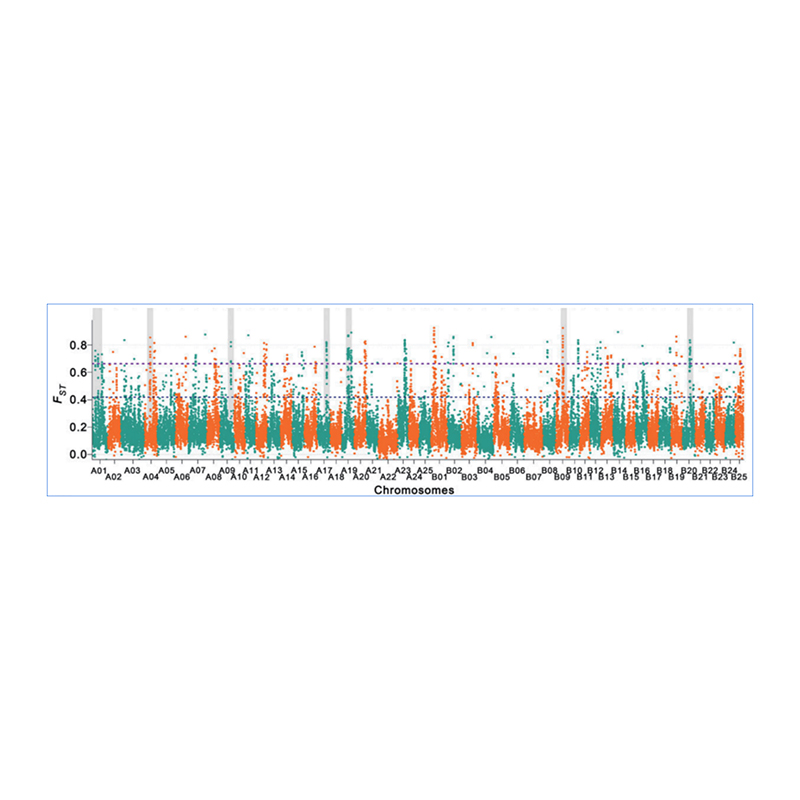
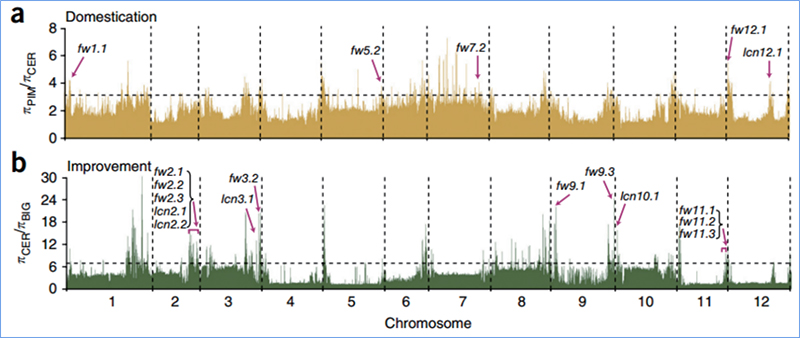
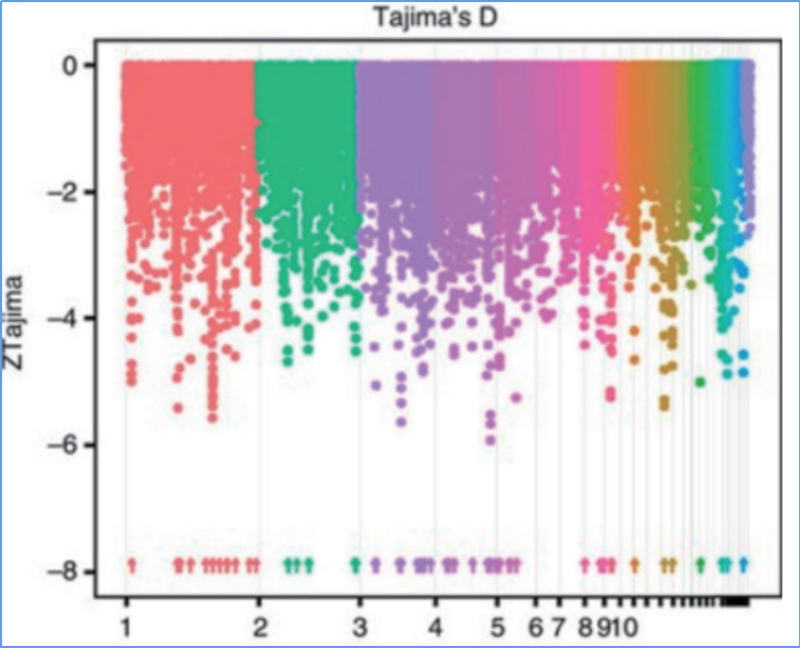
2.Selective sweep
Selective sweep refers to a process by which an advantageous site is selected and frequencies of linked neutral sites are increased and those of unlinked sites are decreased, resulting in reduction of regional.
Genome-wide detection on selective sweep regions is processed by calculating population genetic index(π,Fst, Tajima’s D) of all SNPs within a sliding window (100 Kb) at certain step (10 Kb).
Nucleotide diversity(π)
Tajima’s D
Fixation index(Fst)
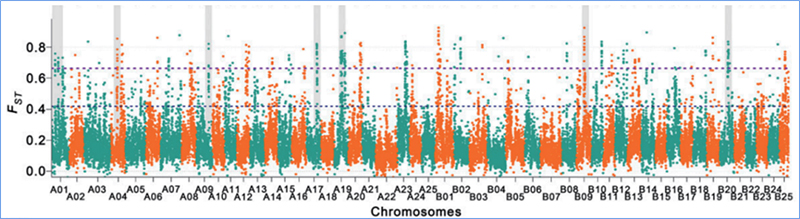
Wu, et. al., Molecular Plant, 2018
3.Gene Flow
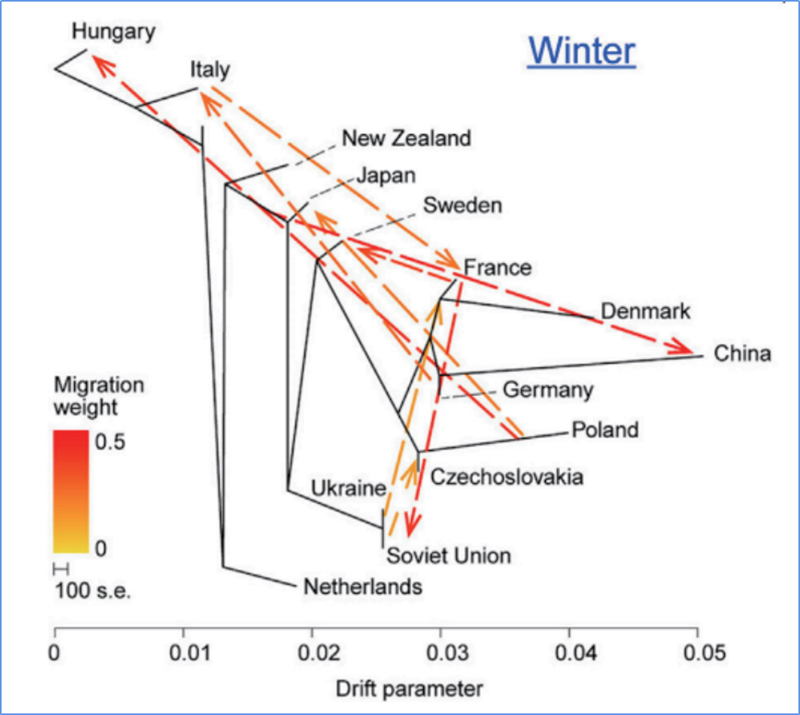
Wu, et. al., Molecular Plant, 2018
4.Demographic history
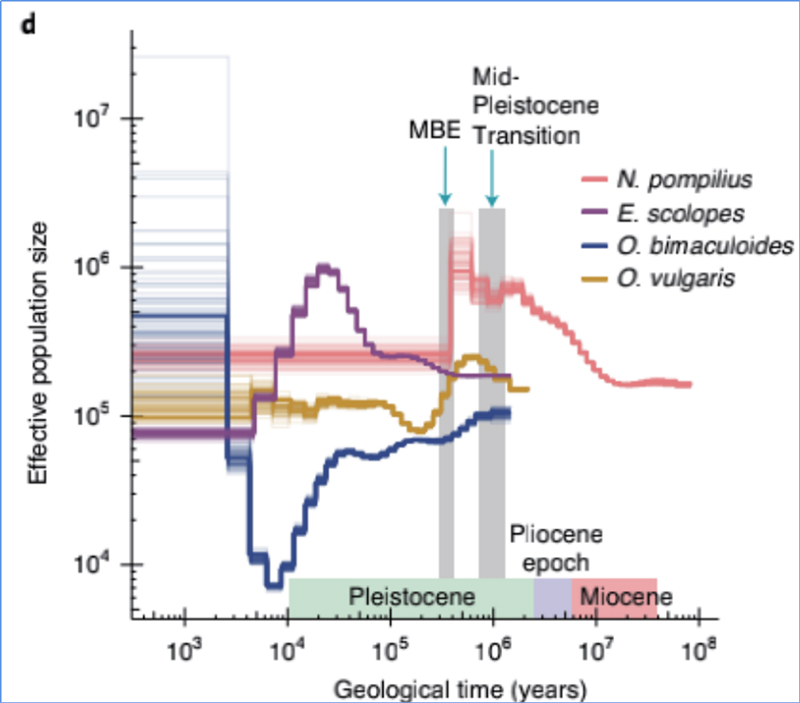
Zhang, et. al., Nature Ecology&Evolution, 2021
5.Divergence time
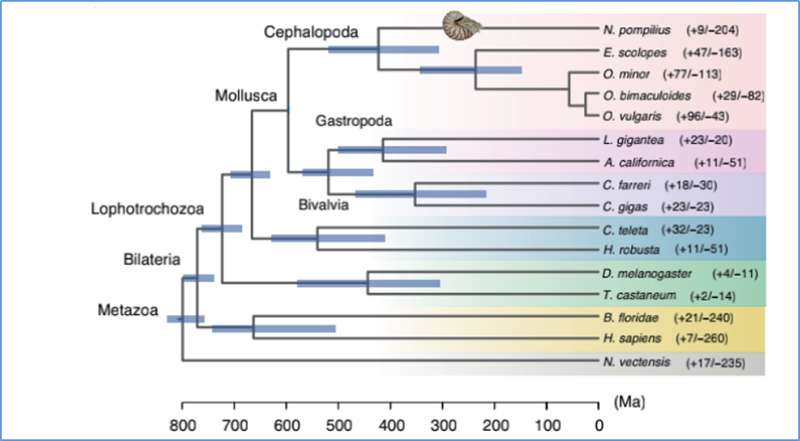
Zhang, et. al., Nature Ecology&Evolution, 2021
Explore the advancements facilitated by BMKGene’s evolutionary genetics services through a curated collection of publications:
Hassanyar, A. K. et al. (2023) ‘Discovery of SNP Molecular Markers and Candidate Genes Associated with Sacbrood Virus Resistance in Apis cerana cerana Larvae by Whole-Genome Resequencing’, International journal of molecular sciences, 24(7). doi: 10.3390/IJMS24076238.
Chai, J. et al. (2022) ‘Discovery of a wild, genetically pure Chinese giant salamander creates new conservation opportunities’, Zoological Research, 2022, Vol. 43, Issue 3, Pages: 469-480, 43(3), pp. 469–480. doi: 10.24272/J.ISSN.2095-8137.2022.101.
Han, M. et al. (2022) ‘Phylogeographical Pattern and Population Evolution History of Indigenous Elymus sibiricus L. on Qinghai-Tibetan Plateau’, Frontiers in Plant Science, 13, p. 882601. doi: 10.3389/FPLS.2022.882601/BIBTEX.
Wang, J. et al. (2022) ‘Genomic insights into longan evolution from a chromosome-level genome assembly and population genomics of longan accessions’, Horticulture Research, 9. doi: 10.1093/HR/UHAC021.