Metagenomic Sequencing -NGS
Service Advantages
● Isolation and cultivation-free method for microbial community profiling: enabling sequencing of genetic material from uncultivable organisms.
● High resolution: in detecting low-abundance species in environmental samples.
● Comprehensive bioinformatics analysis: focused not only on taxonomic diversity but also on the functional diversity of the community.
● Extensive experience: with a track record of successfully closing multiple metagenomics projects in various research domains and processing over 10,000 samples, our team brings a wealth of experience to every project.
Service Specifications
|
Sequencing platform |
Sequencing Strategy |
Data recommended |
Quality control |
|
Illumina NovaSeq or DNBSeq T7 |
PE150 |
6-20Gb |
Q30≥85% |
Service Requirements
|
Concentration (ng/µL) |
Total amount (ng) |
Volume (µL) |
OD260/280 |
|
≥1 |
≥30 |
≥20 |
1.6-2.5 |
● Soil/sludge: 2-3g
● Intestinal content-animal: 0.5-2g
● Intestinal contents-insect: 0.1-0.25g
● Plant surface (enriched sediment): 0.5-1g
● Fermentation broth enriched sediment): 0.2-0.5g
● Faeces (large animals): 0.5-2g
● Faeces (mouse): 3-5grains
● Pulmonary alveolar lavage fluid: filter paper
● Vaginal swab: 5-6 swabs
● Skin/genital swab/saliva/oral soft tissue/pharyngeal swab/rectal swab: 2-3 swabs
● Surface microorganism: 5-6 swabs
● Waterbody/air/biofilm: filter paper
● Endophytes: 2-3g
● Dental Plaque: 0.5-1g
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
Includes the following analysis:
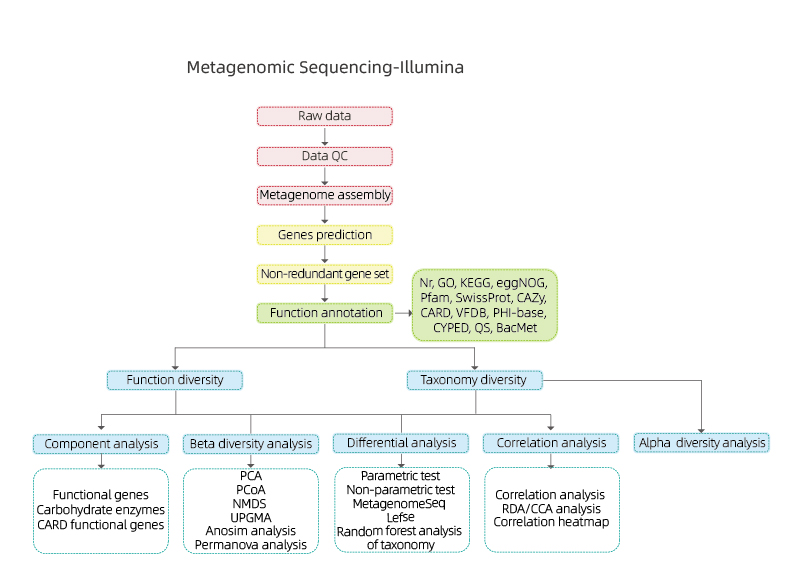
● Sequencing data quality control
● Metagenome assembly and gene prediction
● Gene annotation
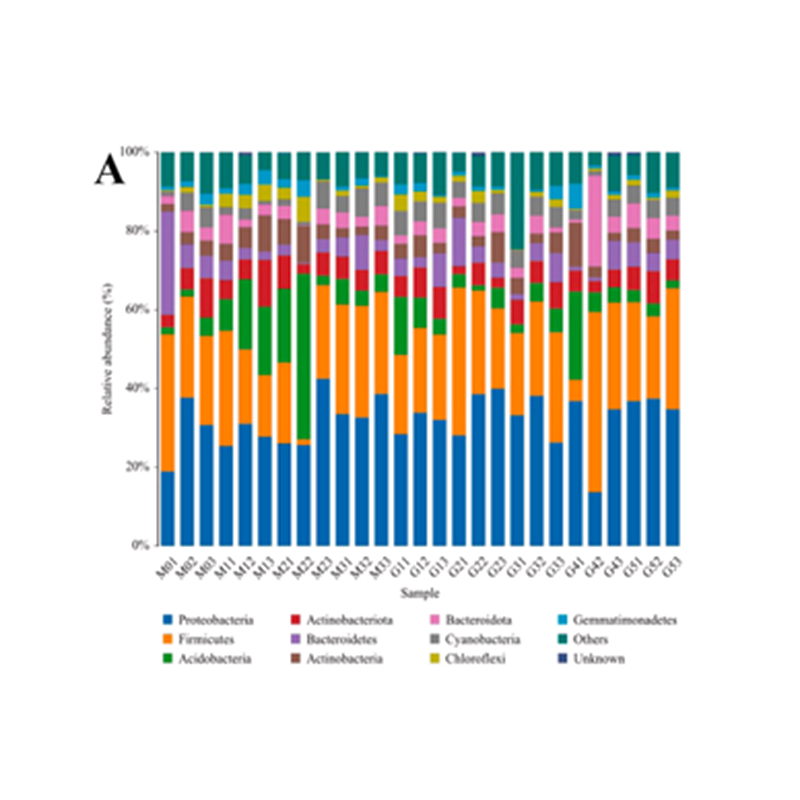
● Taxonomic alpha diversity analysis
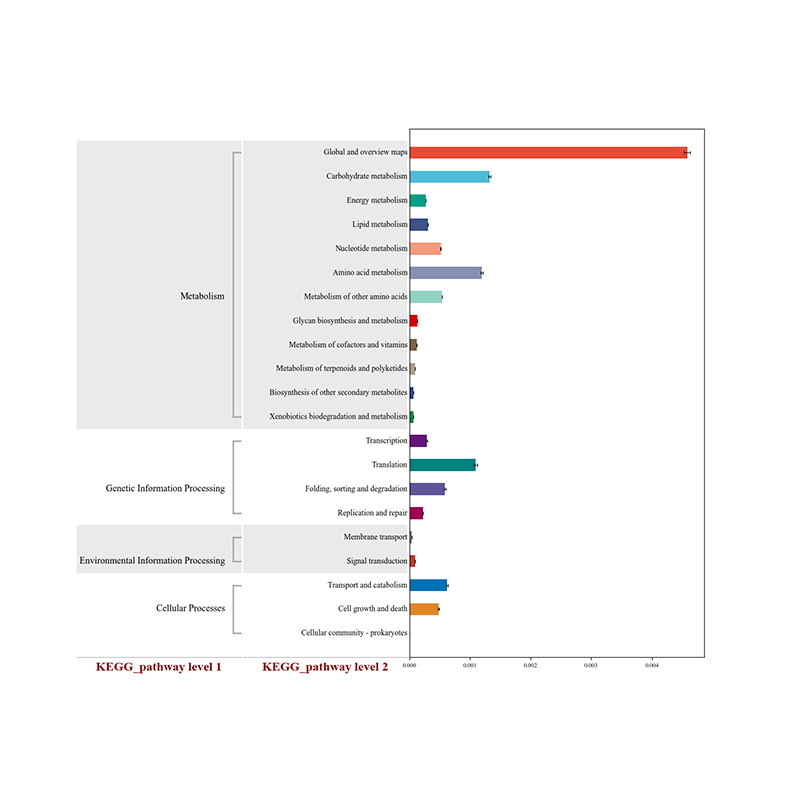
● Functional analysis of the community: biological function, metabolic, antibiotic resistance
● Analysis on both functional and taxonomic diversity:
Beta diversity analysis
Inter-group analysis
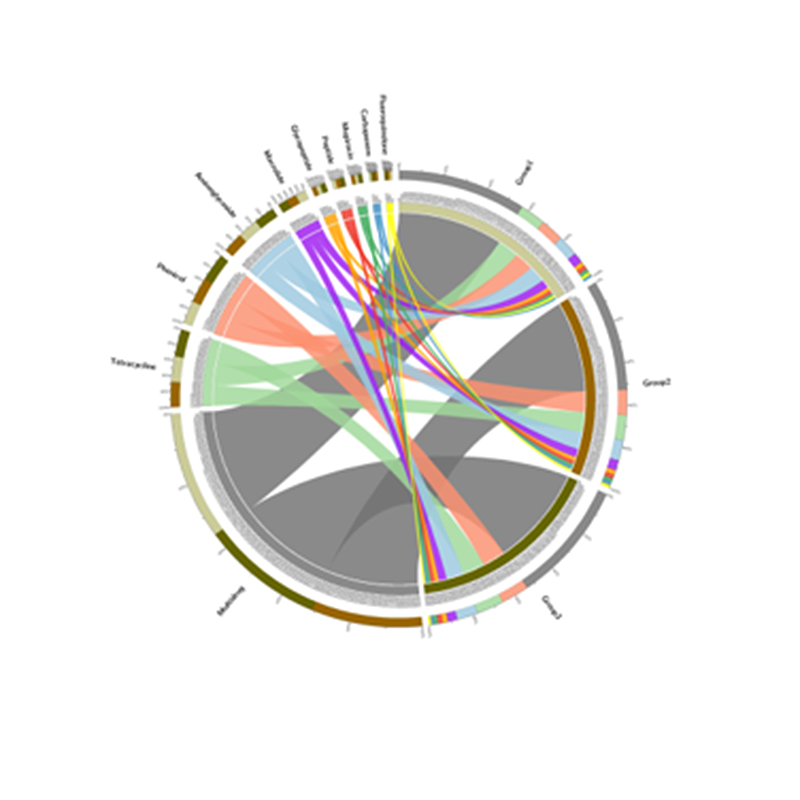
Correlation analysis: between environmental factors and OUT composition and diversity
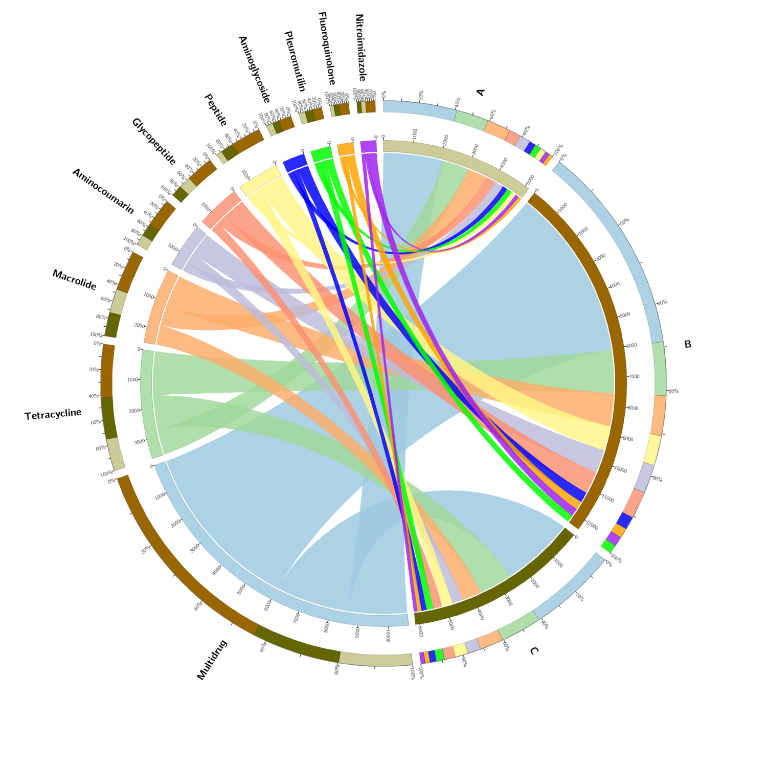
Functional analysis: CARD antibiotic resistance
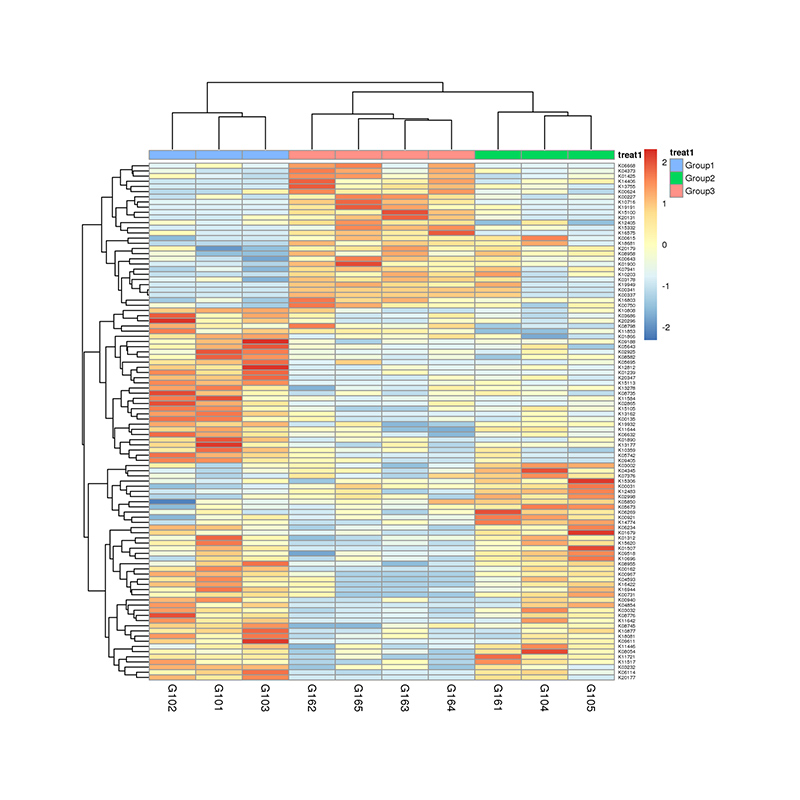
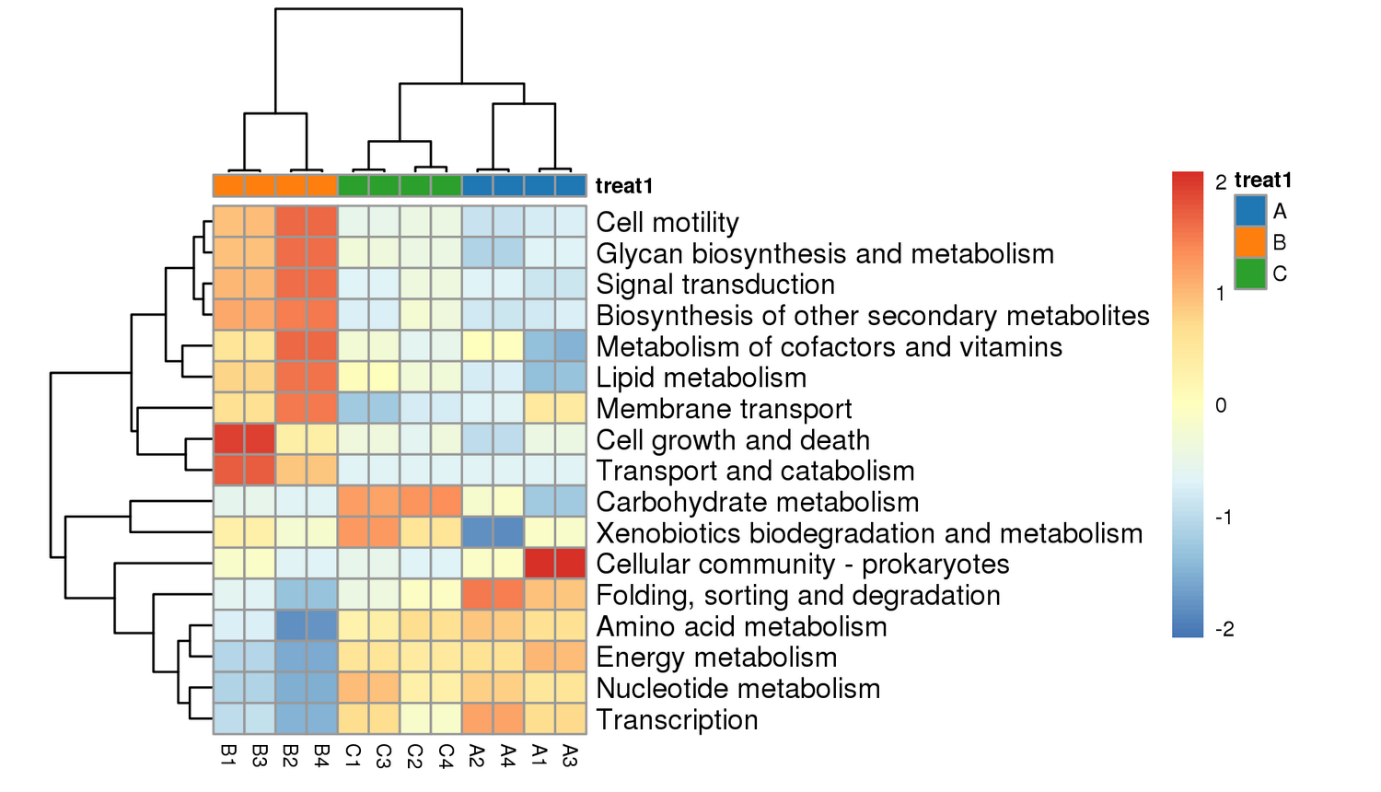
Differential analysis of KEGG metabolic pathways: heatmap of significant pathways
Alpha diversity of taxonomic distribution: ACE index
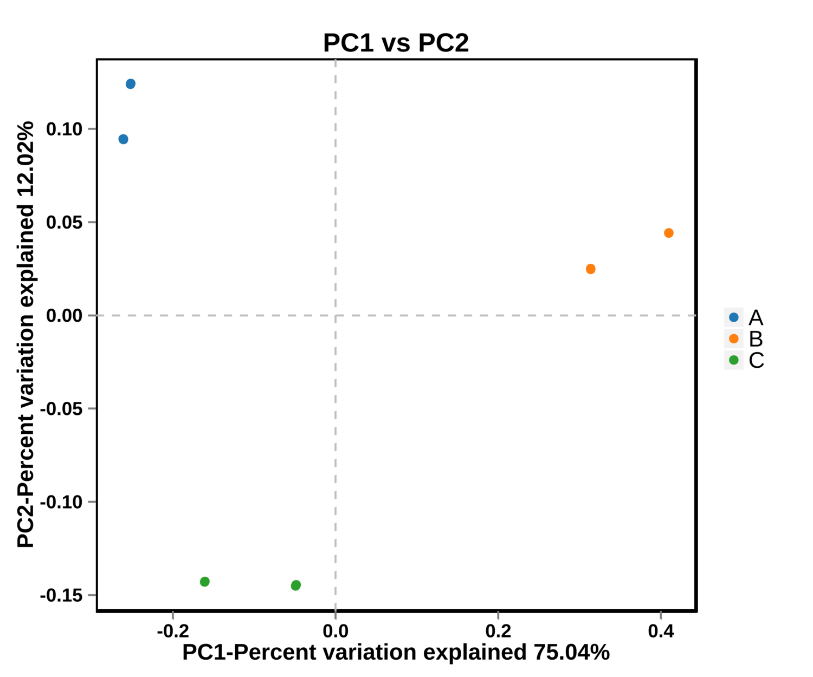
Beta diversity of taxonomic distribution: PCoA
Explore the advancements facilitated by BMKGene’s metagenome sequencing services with Illumina through a curated collection of publications.
Hai, Q. et al. (2023) ‘Metagenomic and metabolomic analysis of changes in intestinal contents of rainbow trout (Oncorhynchus mykiss) infected with infectious hematopoietic necrosis virus at different culture water temperatures’, Frontiers in Microbiology, 14, p. 1275649. doi: 10.3389/FMICB.2023.1275649.
Mao, C. et al. (2023) ‘Microbial communities, resistance genes, and resistome risks in urban lakes of different trophic states: Internal links and external influences’, Journal of Hazardous Materials Advances, 9, p. 100233. doi: 10.1016/J.HAZADV.2023.100233.
Su, M. et al. (2022) ‘Metagenomic Analysis Revealed Differences in Composition and Function Between Liquid-Associated and Solid-Associated Microorganisms of Sheep Rumen’, Frontiers in Microbiology, 13, p. 851567. doi: 10.3389/FMICB.2022.851567.
Yin, J. et al. (2023) ‘Obese Ningxiang pig-derived microbiota rewires carnitine metabolism to promote muscle fatty acid deposition in lean DLY pigs’, The Innovation, 4(5), p. 100486. doi: 10.1016/J.XINN.2023.100486.
Zhao, X. et al. (2023) ‘Metagenomic insights into the potential risks of representative bio/non-degradable plastic and non-plastic debris in the upper and lower reaches of Haihe Estuary, China’, Science of The Total Environment, 887, p. 164026. doi: 10.1016/J.SCITOTENV.2023.164026.