Reduced Representation Bisulfite Sequencing (RRBS)
Service Features
● Requires a reference genome.
● Lambda DNA is used to monitor bisulfite conversion efficiency.
● MspI digestion efficiency is also monitored.
● Sequencing on Illumina NovaSeq.
Service Advantages
● Cost-effective and efficient alternative to WGBS: enabling the analysis the be carried out at a lower cost and with lower sample requirements.
● Complete platform: provide one-stop excellent service from sample processing, library construction, and sequencing to bioinformatics analysis.
● Extensive Expertise: with RRBS sequencing projects successfully completed across a diverse range of species, BMKGENE brings over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
● Possibility to join with transcriptomics analysis: allowing for the integrated analysis of WGBS with other omics data such as RNA-seq.
Service Specifications
|
Library |
Sequencing Strategy |
Recommended data output |
Quality control |
|
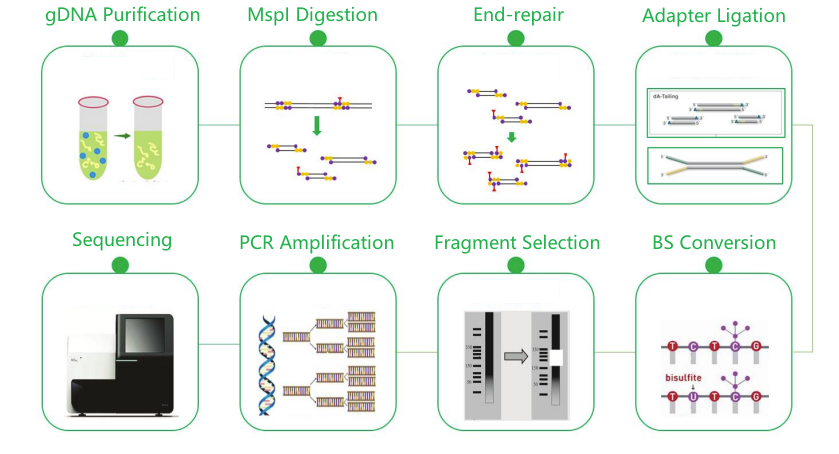
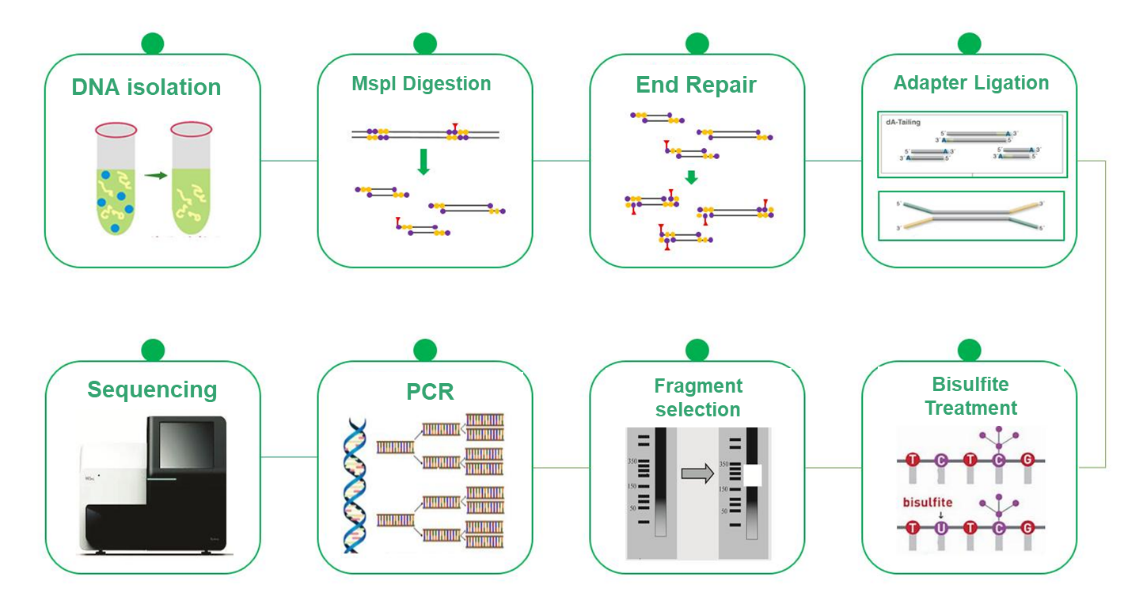
MspI digested and Bisulfite treated library |
Illumina PE150 |
10Gb |
Q30≥85% Bisulfite conversion >99% MspI cutting efficiency > 95% |
Sample Requirements
|
Concentration (ng/µL) |
Total amount (µg) |
|
|
|
Genomic DNA |
≥30 |
≥1 |
Limited degradation or contamination |
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis
After-sale services
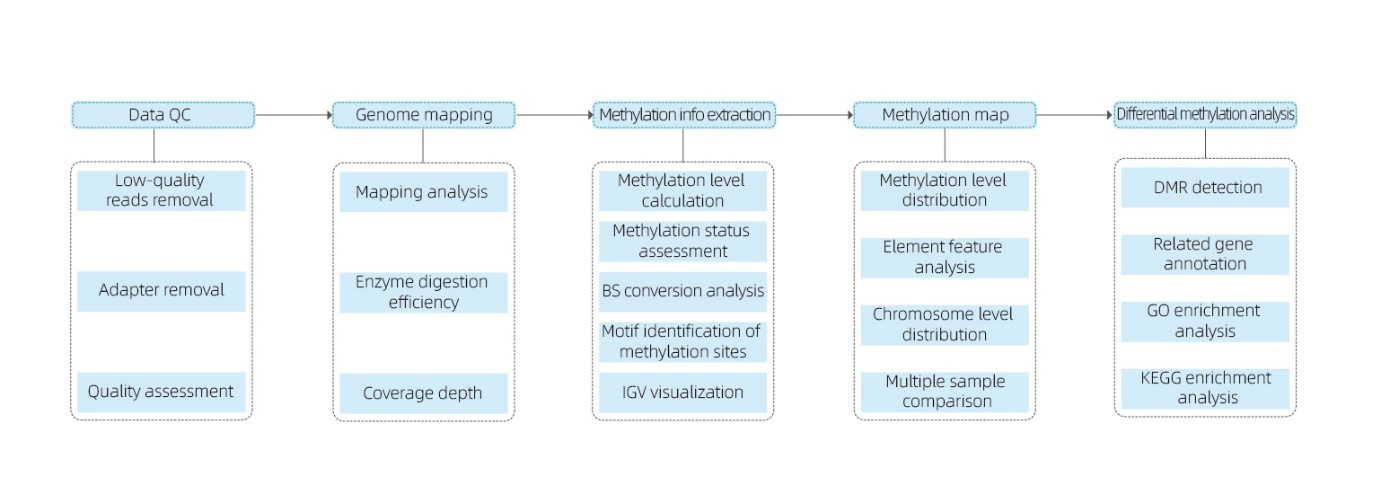
Includes the following analysis:
● Raw sequencing quality control;
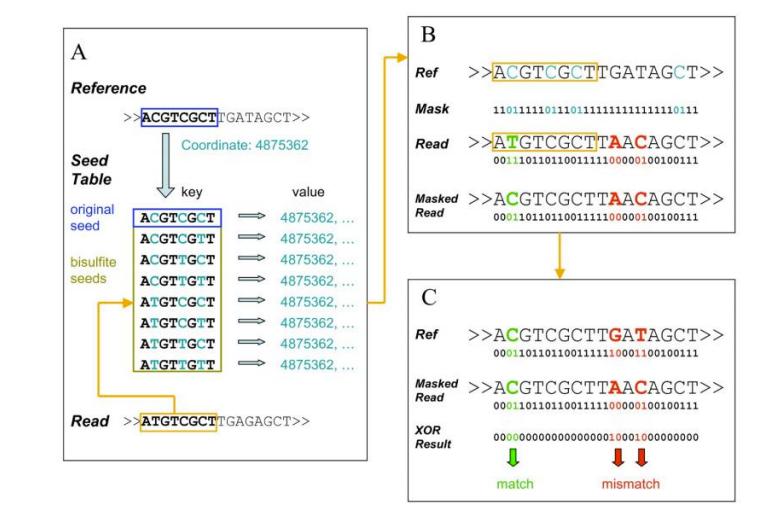
● Mapping to reference genome;
● Detection of 5mC methylated bases and motif identification;
● Analysis of methylation distribution and sample comparison;
● Analysis of Differentially Methylated Regions (DMRs);
● Functional annotation of genes associated to DMRs.
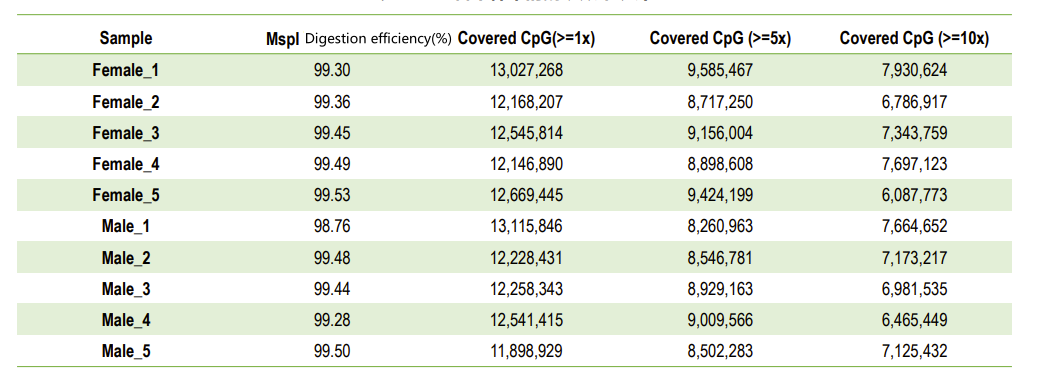
Quality control: digestion efficiency (in genome mapping)
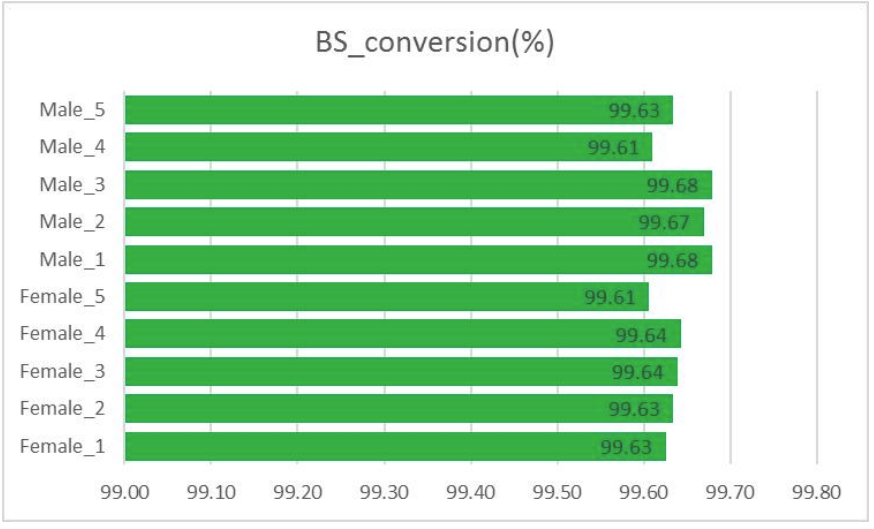
Quality control: bisulfite conversion (in methylation info extraction)
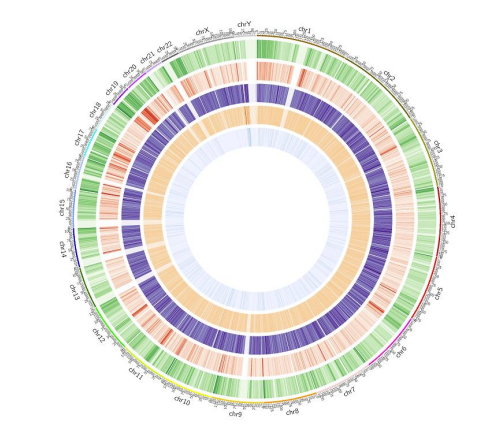
Methylation map: 5mC methylation genome-wide distribution
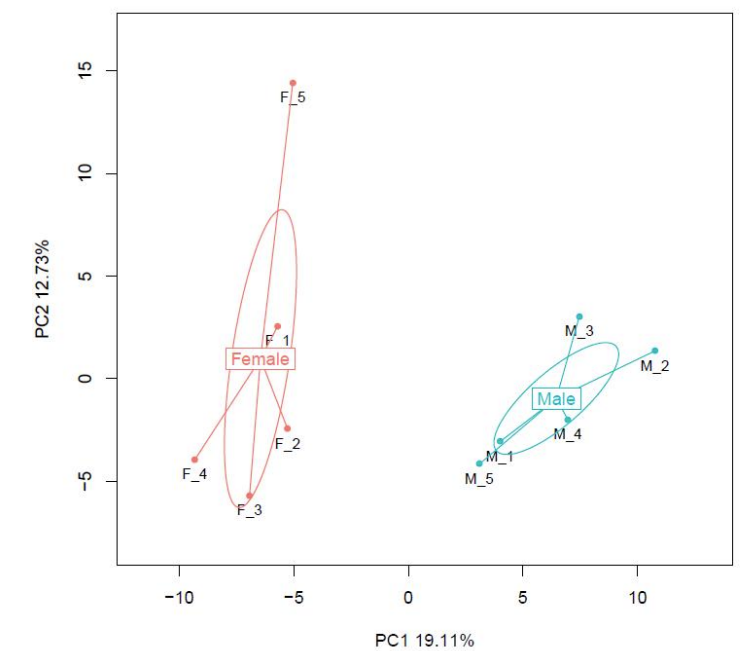
Sample comparison: Principal Component Analysis
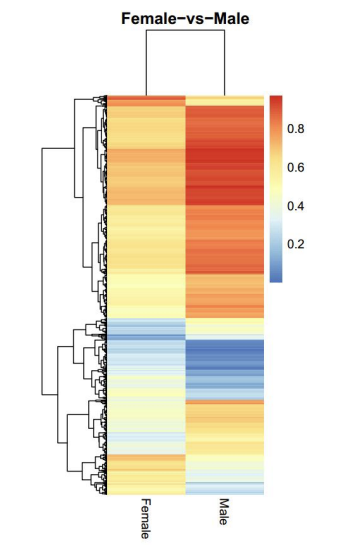
Differentially Methylated Regions (DMRs) analysis: heatmap
Explore the research advancements facilitated by BMKGene’s whole genome bisulfite sequencing services through a curated collection of publications.
Li, Z. et al. (2022) ‘High-fidelity reprogramming into Leydig-like cells by CRISPR activation and paracrine factors’, PNAS Nexus, 1(4). doi: 10.1093/PNASNEXUS/PGAC179.
Tian, H. et al. (2023) ‘Genome-wide DNA methylation analysis of body composition in Chinese monozygotic twins’, European Journal of Clinical Investigation, 53(11), p. e14055. doi: 10.1111/ECI.14055.
Wu, Y. et al. (2022) ‘DNA methylation and waist-to-hip ratio: an epigenome-wide association study in Chinese monozygotic twins’, Journal of Endocrinological Investigation, 45(12), pp. 2365–2376. doi: 10.1007/S40618-022-01878-4.