Genome-wide Association Analysis
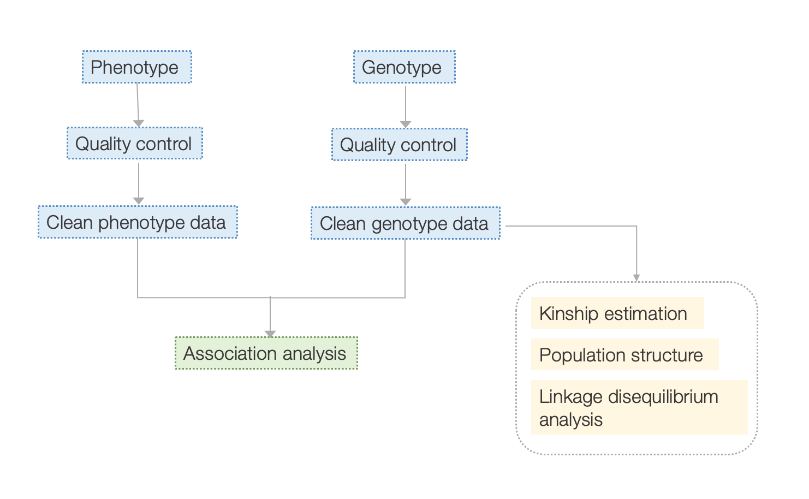
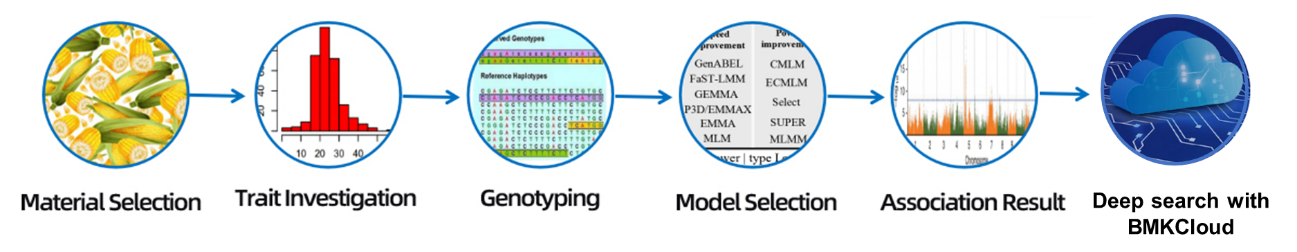
Workflow
Service Advantages
● Extensive Expertise and publication records: with accumulated experience in GWAS, BMKGene has completed hundreds of species projects in population GWAS research, assisted researchers to publish more than 100 articles, and the cumulative impact factor reached 500.
● Comprehensive bioinformatics analysis: workflow includes SNP-trait association analysis, delivering a set of candidate genes and their corresponding functional annotation.
● Highly skilled bioinformatics team and short analysis cycle: with great experience in advanced genomics analysis, BMKGene’s team delivers comprehensive analyses with a swift turnaround time.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Service specifications and requirements
|
Type of sequencing |
Recommended population scale |
Sequencing strategy |
Nucleotide requirements |
|
Whole Genome Sequencing |
200 samples |
10x |
Concentration: ≥1 ng/ µL Total amount≥ 30ng Limited or no degradation or contamination |
|
Specific-Locus Amplified Fragment (SLAF) |
Tag depth: 10x Number of tags: <400 Mb: WGS is recommended <1Gb: 100K tags 1Gb<genome<2Gb: 200K tags >2Gb: 300K tags Max 500k tags |
Concentration ≥ 5 ng/µL Total amount ≥80 ng Nanodrop OD260/280=1.6-2.5 Agarose gel: no or limited degradation or contamination
|
Material Selection



Different varieties, subspecies, landraces/genebanks/mixed families/wild resources
Different varieties, subspecies, landraces
Half-sib family/full-sib family/wild resources
Service Work Flow

Experiment design

Sample delivery

RNA extraction

Library construction

Sequencing

Data analysis

After-sale services
Includes the following analysis:
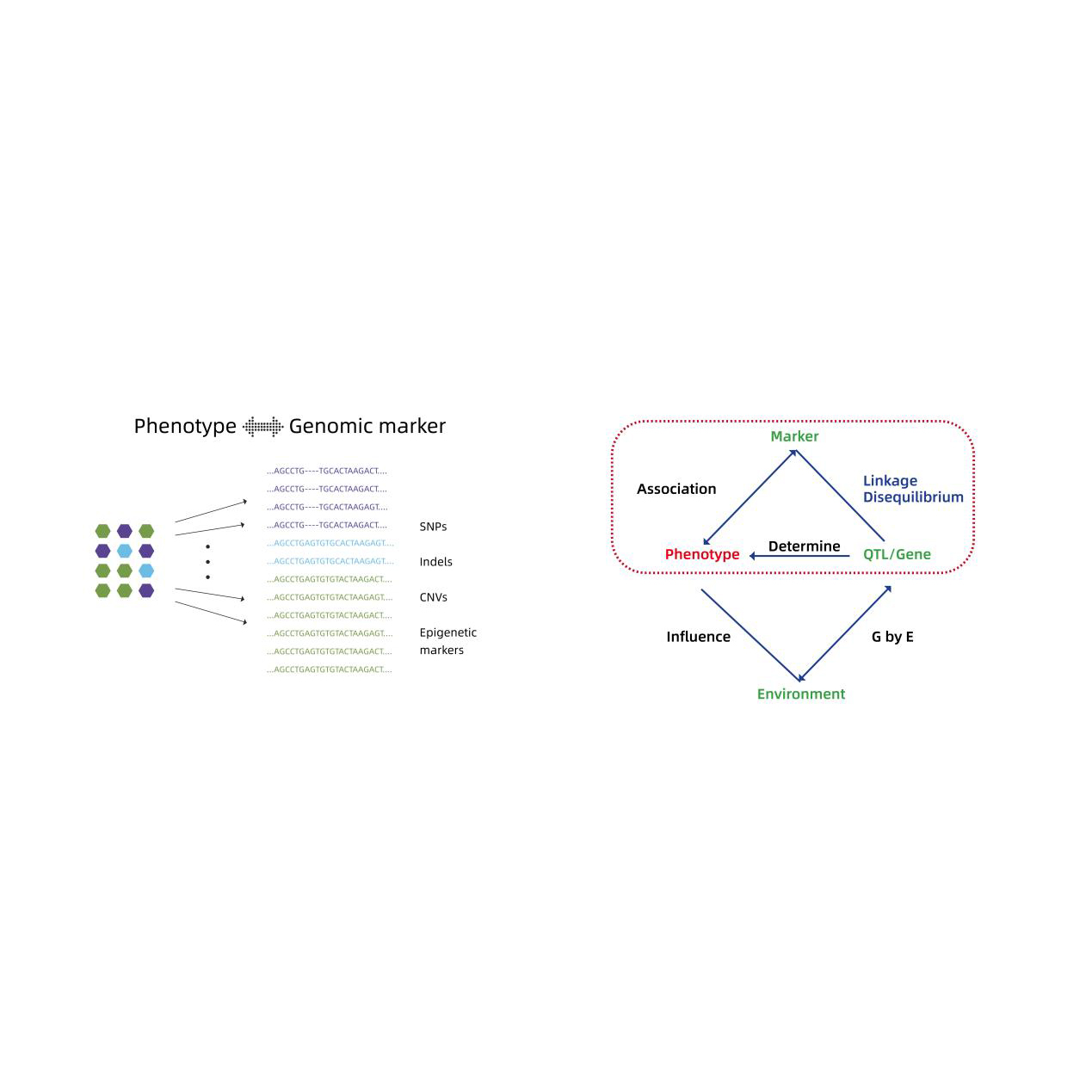
- Genome-wide association analysis: LM, LMM, EMMAX, FASTLMM model
- Functional annotation of candidate genes
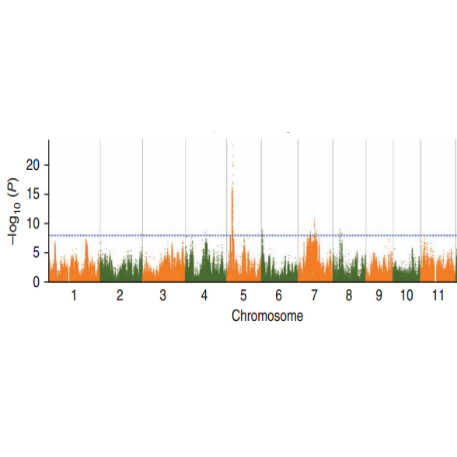
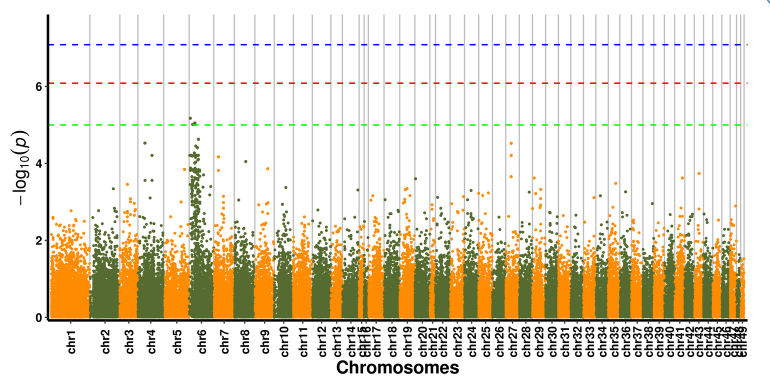
SNP-trait association analysis – Manhattan plot
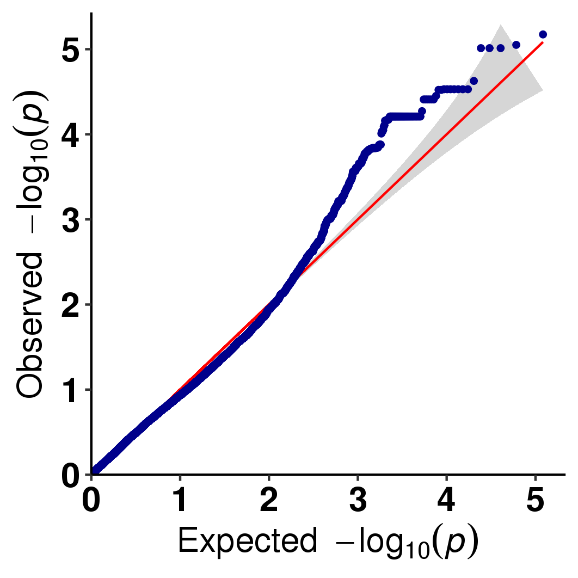
SNP-trait association analysis – QQ plot
Explore the advancements facilitated by BMKGene’s de GWAS services through a curated collection of publications:
Lv, L. et al. (2023) ‘Insight into the genetic basis of ammonia tolerance in razor clam Sinonovacula constricta by genome-wide association study’, Aquaculture, 569, p. 739351. doi: 10.1016/J.AQUACULTURE.2023.739351.
Li, X. et al. (2022) ‘Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti-inflammatory effects’, Molecular Plant, 15(8), pp. 1367–1383. doi: 10.1016/j.molp.2022.07.003.
Li, J. et al. (2022) ‘Genome-Wide Association Mapping of Hulless Barely Phenotypes in Drought Environment’, Frontiers in Plant Science, 13, p. 924892. doi: 10.3389/FPLS.2022.924892/BIBTEX.
Zhao, X. et al. (2021) ‘GmST1, which encodes a sulfotransferase, confers resistance to soybean mosaic virus strains G2 and G3’, Plant, Cell & Environment, 44(8), pp. 2777–2792. doi: 10.1111/PCE.14066.