16S/18S/ITS Amplicon Sequencing-PacBio
Service Features
● Sequencing platform: PacBio Revio
● Sequencing mode: CCS (HiFi reads)
● Amplification of target region followed by tandem linking of amplicons before HiFi SMRT bell library preparation
Service Advantages
● Higher taxonomic resolution: than short-amplicon sequencing, enabling higher OTU classification rates at the species level.
● Highly accurate base calling: with PacBio CCS mode sequencing (HiFi reads).
● Isolation-free: and rapid identification of microbial composition in environmental samples.
● Widely applicable: to diverse microbial community studies.
● Comprehensive bioinformatic analysis: including with the latest QIIME2 package (quantitative insight into microbial ecology) with diverse analyses in terms of database, annotation, OTU/ASV.
● Extensive Expertise: with thousands of amplicon sequencing projects conducted annually, BMKGENE brings over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
Service Specifications
|
Library |
Sequencing Strategy |
Data recommended |
Quality control |
|
Amplicon |
PacBio Revio |
10/20 K tags (read pairs) |
Q30≥85% |
Sample Requirements
|
Concentration (ng/µL) |
Total amount (µg) |
Volume (µL) |
OD260/280 |
|
≥20 |
≥1 |
≥20 |
1.6-2.5 |
Recommended Sample Delivery
Freeze the samples in liquid nitrogen for 3-4 hours and store in liquid nitrogen or -80 degree to long-term reservation. Sample shipping with dry-ice is required.
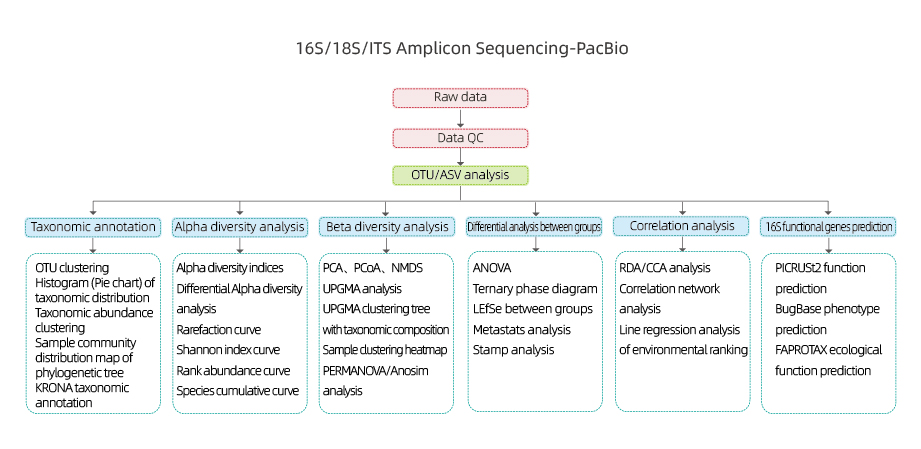
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
Includes the following analysis:
●Raw data quality control
●OTU clustering/De-noise(ASV)
●OTU annotation
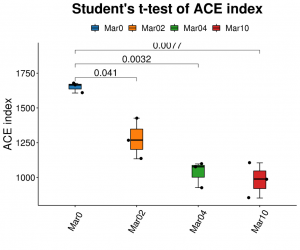
●Alpha diversity analysis: multiple indexes, including Shannon, Simpson and ACE.
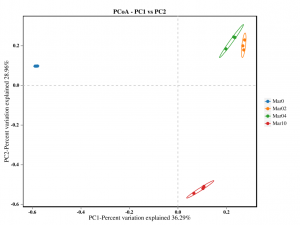
●Beta diversity analysis
●Inter-group analysis
●Correlation analysis: between environmental factors and OUT composition and diversity
●16S functional gene prediction
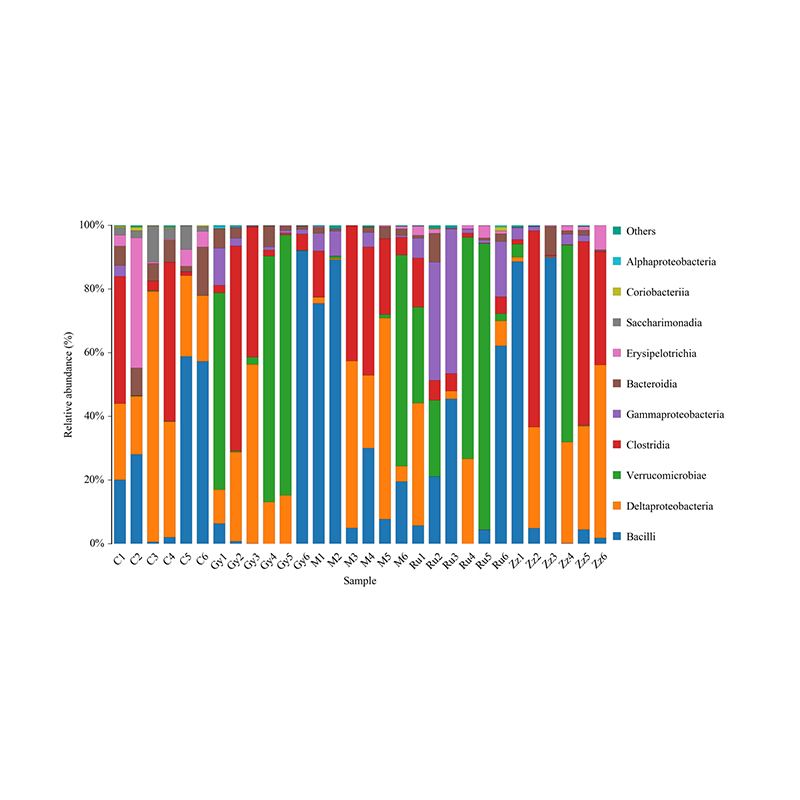
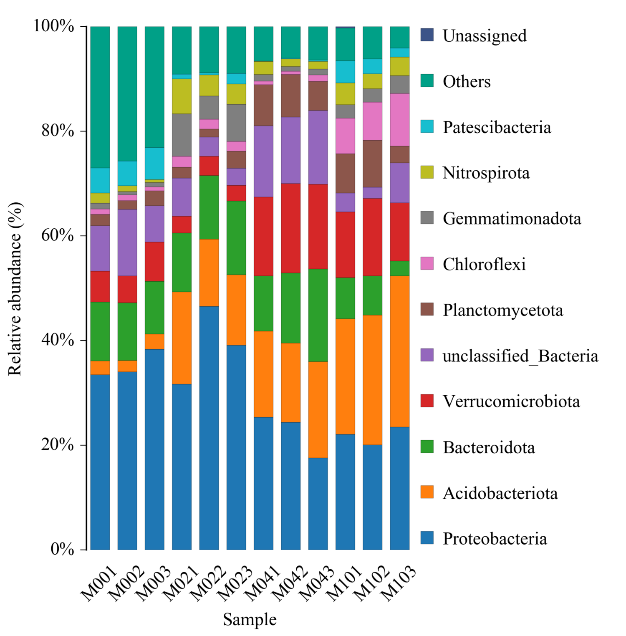
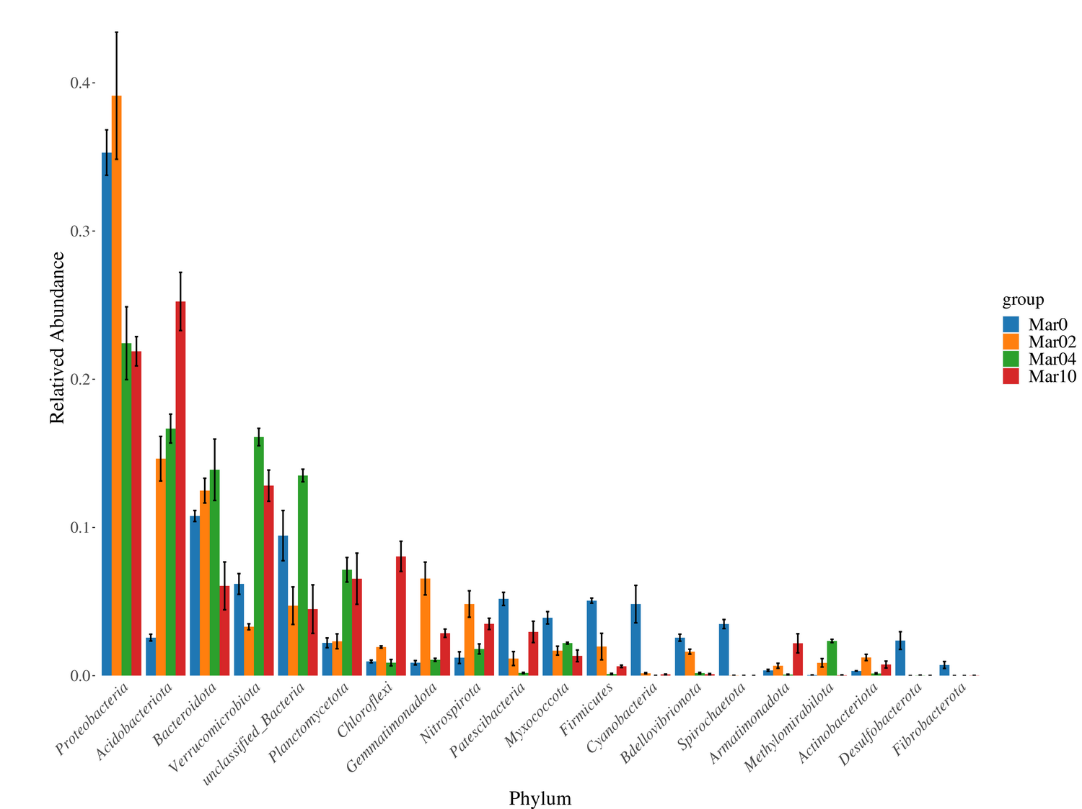
Histogram of taxonomic distribution
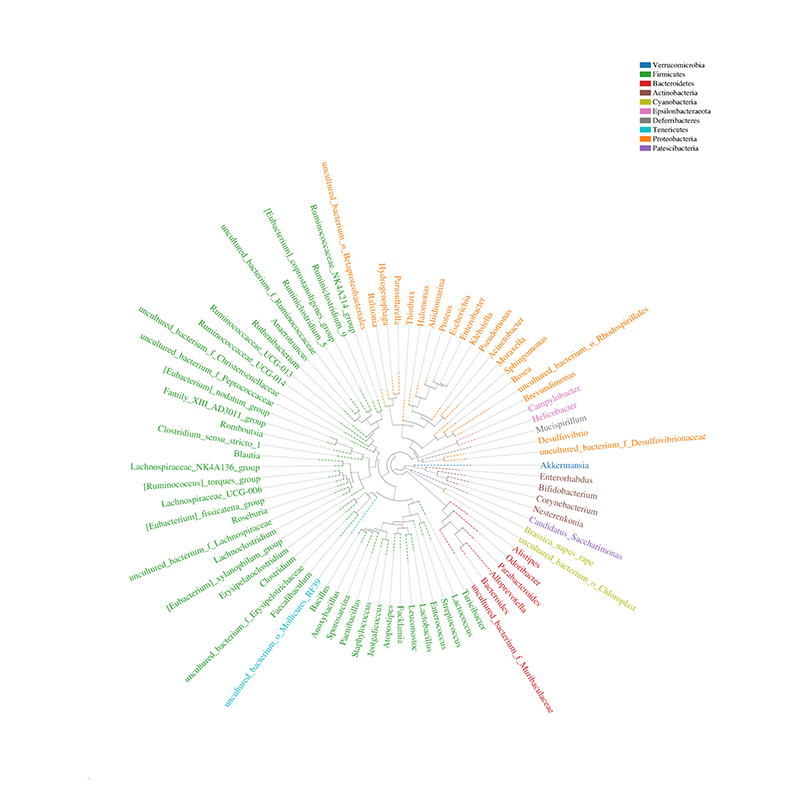
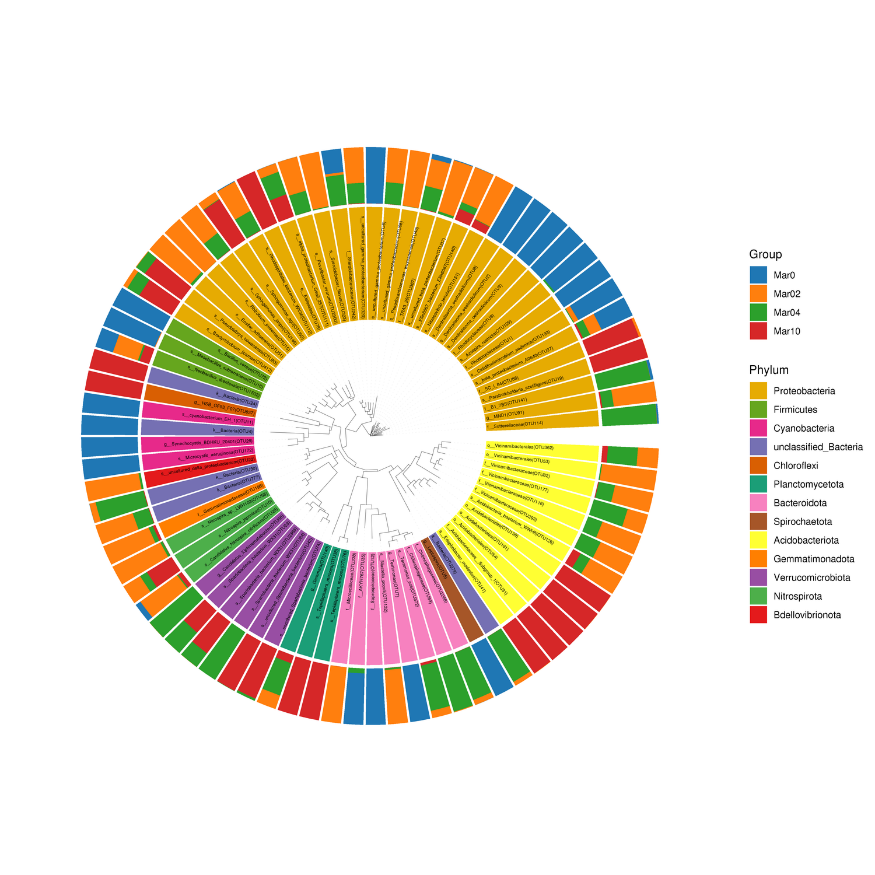
Community distribution phylogenetic tree
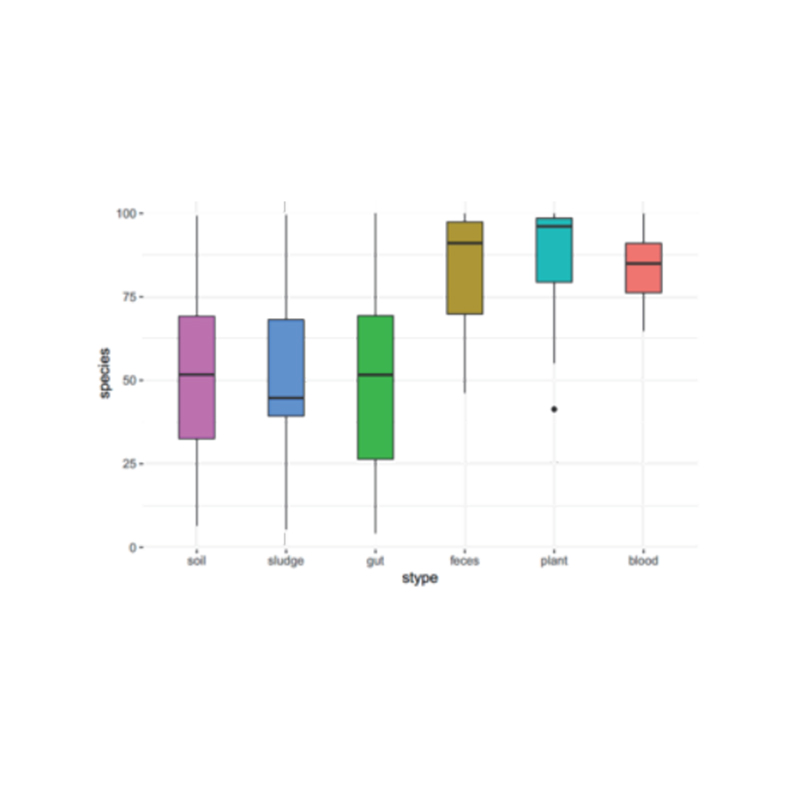
Alpha diversity analysis: ACE
Beta diversity analysis: PCoA
Intergroup analysis: ANOVA
Explore the advancements facilitated by BMKGene’s amplicon sequencing services with PacBio through a curated collection of publications.
Gao, X. and Wang, H. (2023) ‘Comparative Analysis of Rumen Bacterial Profiles and Functions during Adaption to Different Phenology (Regreen vs. Grassy) in Alpine Merino Sheep with Two Growing Stages on an Alpine Meadow’, Fermentation, 9(1), p. 16. doi: 10.3390/FERMENTATION9010016/S1.
Li, S. et al. (2023) ‘Capturing the microbial dark matter in desert soils using culturomics-based metagenomics and high-resolution analysis’, npj Biofilms and Microbiomes 2023 9:1, 9(1), pp. 1–14. doi: 10.1038/s41522-023-00439-8.
Mu, L. et al. (2022) ‘Effects of fatty acid salts on fermentation characteristics, bacterial diversity and aerobic stability of mixed silage prepared with alfalfa, rice straw and wheat bran’, Journal of the Science of Food and Agriculture, 102(4), pp. 1475–1487. doi: 10.1002/JSFA.11482.
Yang, J. et al. (2023) ‘The Interaction between Oxidative Stress Biomarkers and Gut Microbiota in the Antioxidant Effects of Extracts from Sonchus brachyotus DC. in Oxazolone-Induced Intestinal Oxidative Stress in Adult Zebrafish’, Antioxidants 2023, Vol. 12, Page 192, 12(1), p. 192. doi: 10.3390/ANTIOX12010192.