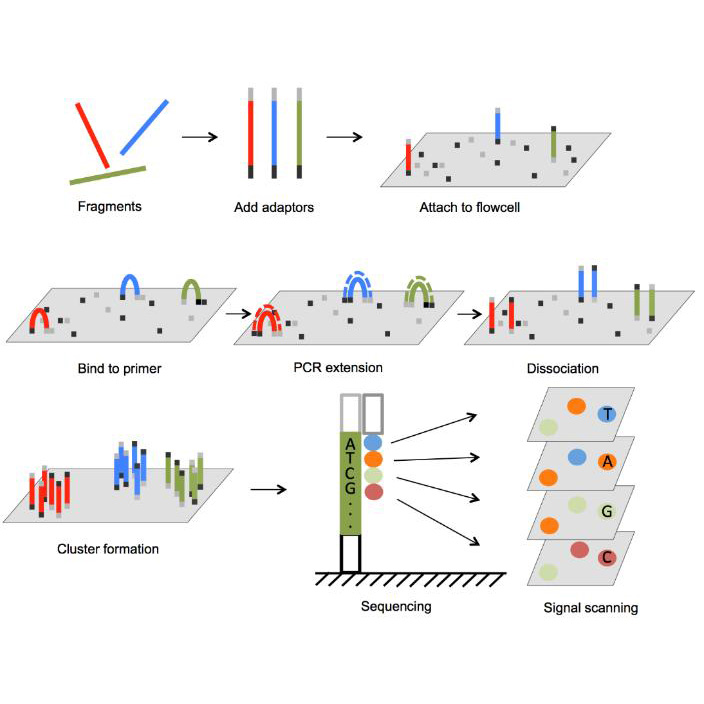
Illumina pre-made libraries
Features
● Platforms: Illumina NovaSeq 6000 and NovaSeq X
● Sequencing modes: PE150 and PE250
● Quality control of libraries before sequencing
● Sequencing data delivery and QC: delivery of QC report and raw data in fastq format after demultiplexing and filtering Q30 reads.
Service Advantages
● Versatility of Sequencing services: the customer may choose to sequence by lane, flow cell, or by amount of data required (partial lane sequencing).
● Extensive experience on Illumina sequencing platform: with thousands of closed projects with various species.
● Delivery of sequencing QC report: with quality metrics, data accuracy and overall performance of the sequencing project.
● Mature sequencing process: with short turn-around time.
● Rigorous Quality Control: we implement strict QC requirements to guarantee the delivery of consistently high-quality results.
Sample Platforms
|
Platform |
Flow Cell |
Sequencing mode |
Unit |
Estimated output |
|
NovaSeq X |
10B (8 lanes) |
PE150 |
Single Lane Partial Lane |
375Gb /Lane |
|
25B ( lanes) |
PE150 |
Single Lane Partial Lane |
1000 Gb/Lane |
|
|
NovaSeq 6000 |
SP Flow cell (2 lanes) |
PE250 |
Flow Cell Single Lane Partial Lane |
~162-200Gb/ Lane |
|
S4 Flow cell (4 lanes) |
PE150 |
Flow Cell Single Lane Partial Lane |
~800 Gb/Lane |
Sample Requirements
|
Data Amount (X) /Platform |
Concentration (qPCR/nM) |
Volume |
|
|
Partial Lane Sequencing |
X ≤ 50 Gb |
≥ 2 nM |
≥ 20 μl |
|
50 Gb ≤ X < 100 Gb |
≥ 3 nM |
≥ 20 μl |
|
|
X ≥ 100 Gb |
≥ 4 nM |
≥ 20 μl |
|
|
Lane Sequencing |
NovaSeq 6000 SP |
≥ 1 nM |
≥ 25 μl |
|
NovaSeq 6000 S4 |
≥ 1.5 nM |
≥ 25 μl |
|
|
NovaSeq X |
≥ 1.5 nM |
≥ 25 μl |
In addition to concentration and total amount, a suitable peak pattern is also required.
Please reach out to us if your samples don't meet the starting material requirements.
Service Workflow
Library quality control

Sequencing

Data quality control

Project delivery
Library QC report
A report on the quality of the library is provided before sequencing, assessing library amount, and fragmentation.
Sequencing QC report
Table 1. Statistics on sequencing data.
|
Sample ID |
BMKID |
Raw reads |
Raw Data (bp) |
Clean reads (%) |
Q20(%) |
Q30(%) |
GC(%) |
|
C_01 |
BMK_01 |
22,870,120 |
6,861,036,000 |
96.48 |
99.14 |
94.85 |
36.67 |
|
C_02 |
BMK_02 |
14,717,867 |
4,415,360,100 |
96.00 |
98.95 |
93.89 |
37.08 |
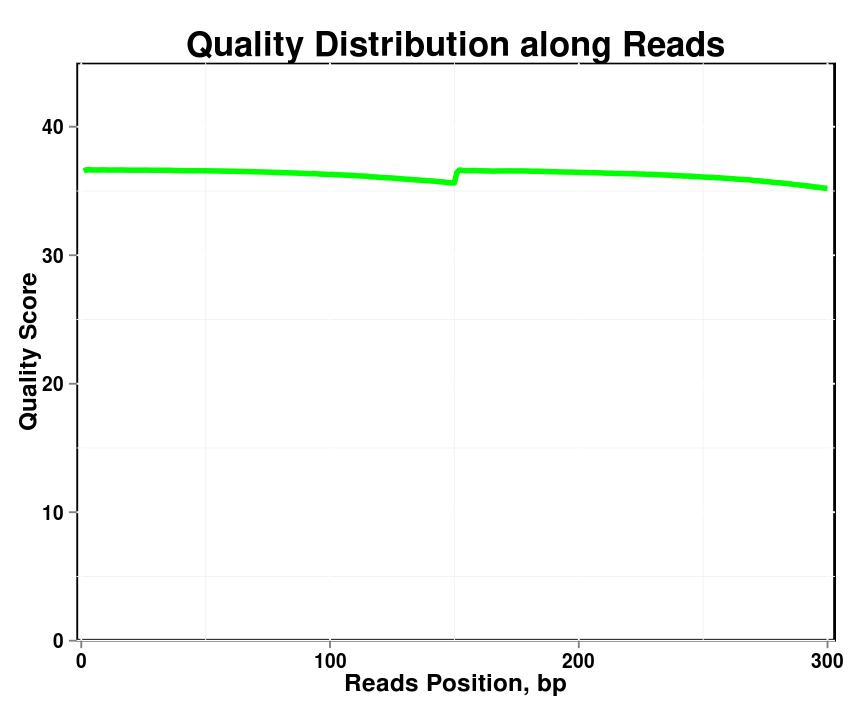
Figure 1. Quality distribution along reads in each sample
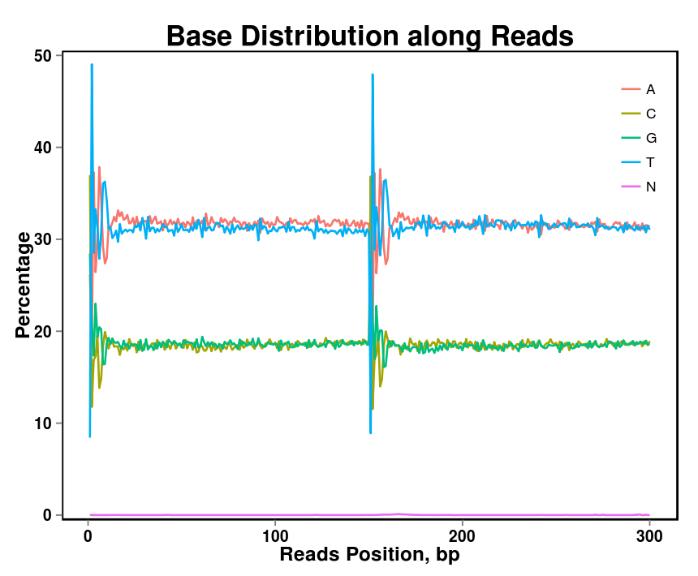
Figure 2. Base content distribution
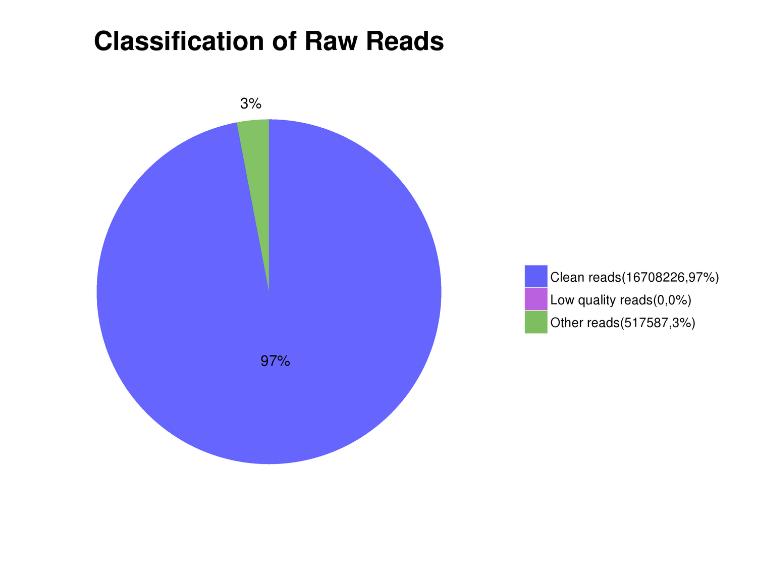
Figure 3. Distribution of read contents in sequencing data