-

Amplicon (16S/18S/ITS)
Amplicon (16S/18S/ITS) platform is developed with years of experience in microbial diversity project analysis, which contains standardized basic analysis and personalized analysis: basic analysis covers the mainstream analysis content of current microbial research, the analysis content is rich and comprehensive, and analysis results are presented in the form of project reports; The content of personalized analysis is diverse. Samples can be selected and parameters can be set flexibly according to the basic analysis report and research purpose, to realize personalized requirements. Windows operating system, simple and fast.
-

Evolutionary Genetics
Evolutionary Genetics is a comprehensive sequencing service designed to offer an insightful interpretation of the evolution within a large group of individuals, based on genetic variations, including SNPs, InDels, SVs, and CNVs. This service encompasses all essential analyses needed to elucidate the evolutionary shifts and genetic characteristics of populations, including assessments of population structure, genetic diversity, and phylogenetic relationships. Moreover, it delves into studies on gene flow, enabling estimations of effective population size and divergence time. Evolutionary genetics studies yield valuable insights into the origins and adaptations of species.
At BMKGene, we offer two avenues for conducting evolutionary genetics studies on large populations: employing whole-genome sequencing (WGS) or opting for a reduced representation genome sequencing method, the in-house-developed Specific-Locus Amplified Fragment (SLAF). While WGS suits smaller genomes, SLAF emerges as a cost-effective alternative for studying larger populations with longer genomes, effectively minimizing sequencing costs.
-

Comparative Genomics
Comparative genomics involves the examination and comparison of the entire genome sequences and structures among different species. This field seeks to unveil the evolution of species, decode gene functions, and elucidate the genetic regulatory mechanisms by identifying conserved or divergent sequence structures and elements across various organisms. A comprehensive comparative genomics study encompasses analyses such as gene families, evolutionary development, whole-genome duplication events, and the impact of selective pressures.
-

Evolutionary Genetics
Population and evolutionary genetic analysis platform is established on base of massive experience accumulated within BMK R&D team for years. It is a user friendly tool especially for researchers who are not majoring in bioinformatics. This platform enables basic evolutionary genetics related basic analysis including phylogenetic tree construction, linkage disequilibrium analysis, genetic diversity assessment, selective sweep analysis, kinship analysis, PCA, population structure analysis, etc.
-
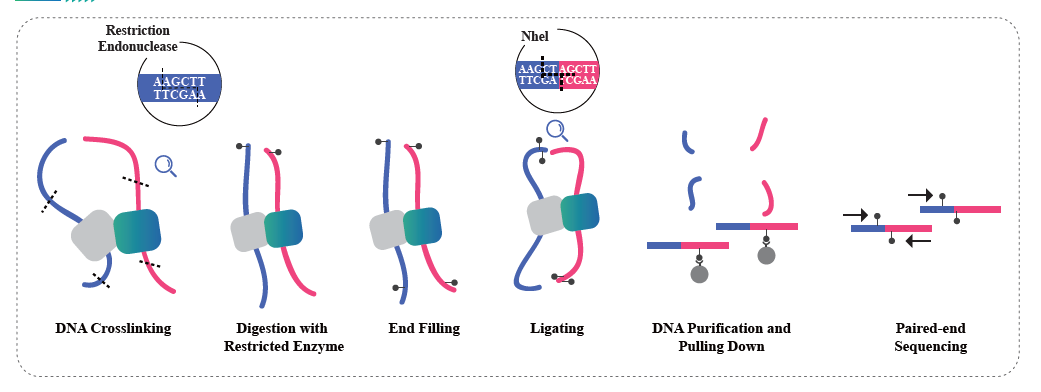
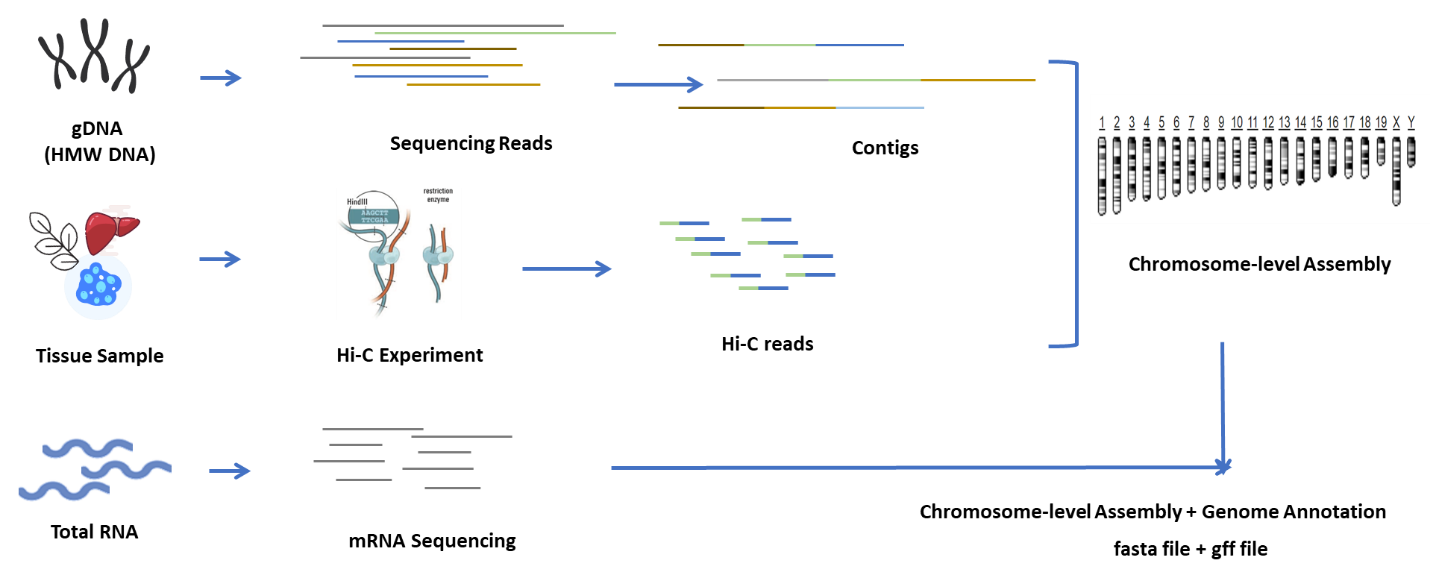
Hi-C based Genome Assembly
Hi-C is a method designed to capture chromosome configuration by combining probing proximity-based interactions and high-throughput sequencing. The intensity of these interactions is believed to be negatively correlated with physical distance on chromosomes. Therefore, Hi-C data is used to guide the clustering, ordering, and orienting of assembled sequences in a draft genome and anchoring those onto a certain number of chromosomes. This technology empowers a chromosome-level genome assembly in the absence of a population-based genetic map. Every single genome needs a Hi-C.
-

Plant/Animal De Novo Genome Sequencing
De Novo sequencing refers to the construction of a species’ whole genome using sequencing technologies in the absence of a reference genome. The introduction and widespread adoption of third-generation sequencing, featuring longer reads, have significantly enhanced genome assembly by increasing the overlap between reads. This enhancement is particularly pertinent when dealing with challenging genomes, such as those exhibiting high heterozygosity, a high ratio of repetitive regions, polyploids, and regions with repetitive elements, abnormal GC contents, or high complexity that are typically poorly assembled using short-read sequencing alone.
Our one-stop solution provides integrated sequencing services and bioinformatic analysis that deliver a high-quality de novo assembled genome. An initial genome survey with Illumina provides estimations of genome size and complexity, and this information is used to guide the next step of long-read sequencing with PacBio HiFi, followed by de novo assembly of contigs. The subsequent use of HiC assembly enables anchoring of the contigs to the genome, obtaining a chromosome-level assembly. Finally, the genome is annotated by gene prediction and by sequencing expressed genes, resorting to transcriptomes with short and long reads.
-

Human Whole Exome Sequencing
Human Whole exome sequencing (hWES) is widely acknowledged as a cost-effective and powerful sequencing approach for pinpointing disease-causing mutations. Despite constituting only about 1.7% of the entire genome, exons play a crucial role by directly reflecting the profile of total protein functions. Notably, in the human genome, over 85% of mutations related to diseases manifest within the protein coding regions. BMKGENE offers a comprehensive and flexible human whole exome sequencing service with two different exon capturing strategies available to meet various research goals.
-

Specific-Locus Amplified Fragment Sequencing (SLAF-Seq)
High-throughput genotyping, particularly on large-scale populations, is fundamental step in genetic association studies and provides a genetic basis for functional gene discovery, evolutionary analysis, etc. Instead of deep whole genome re-sequencing, Reduced Representation Genome Sequencing (RRGS) is often employed in these studies to minimize sequencing cost per sample while maintaining reasonable efficiency in genetic marker discovery. RRGS achieves this by digesting DNA with restriction enzymes and focusing on a specific fragment size range, thereby sequencing only a fraction of the genome. Among the various RRGS methodologies, Specific-Locus Amplified Fragment Sequencing (SLAF) is a customizable and high-quality approach. This method, developed independently by BMKGene, optimizes the restriction enzyme set for every project. This ensures the generation of a substantial number of SLAF tags (400-500 bps regions of the genome being sequenced) that are uniformly distributed across the genome while effectively avoiding repetitive regions, thus assuring the best genetic marker discovery.
-

Illumina pre-made libraries
Illumina sequencing technology, based on Sequencing by Synthesis (SBS), is a globally embraced NGS innovation, responsible for generating over 90% of the world’s sequencing data. The principle of SBS involves imaging fluorescently labelled reversible terminators as each dNTP is added, and subsequently cleaved to allow the incorporation of the next base. With all four reversible terminator-bound dNTPs present in each sequencing cycle, natural competition minimizes incorporation bias. This versatile technology supports both single-read and paired-end libraries, catering to a range of genomic applications. Illumina sequencing’s high-throughput capabilities and precision position it as a cornerstone in genomics research, empowering scientists to unravel the intricacies of genomes with unmatched detail and efficiency.
Our pre-made library sequencing service enables customers to prepare sequencing libraries from diverse sources (mRNA, whole genome, amplicon, 10x libraries, among others). Subsequently, these libraries can be shipped to our sequencing centers for quality control and sequencing in Illumina platforms.