GENOME EVOLUTION
PNAS
The evolutionary origin and domestication history of goldfish (Carassius auratus)
PacBio | Illumina | Bionano Genome Map | Hi-C Genome Assembly | Genetic Map | GWAS | RNA-Seq
Highlights
1.Goldfish genome was updated with a high-quality assembly version, anchoring 95.75% of contigs into 50 pseudochromosomes (Scaffold N50=31.84 Mb). Two subgenomes were disentangled.
2.Genomic regions of selective sweeps during domestication were identified from resequencing data of 201 individuals, unveiling over 390 candidate genes that are likely associated with domestication traits.
3.GWAS on dorsal fin in domesticated goldfish revealed 378 candidate genes that potentially associated. A tyrosine-protein kinase reporter was identified as a candidate causal gene associated with transparency
Background
Goldfish (Carassius auratus) is one of the most important farmed fish, which were domesticated from crucian carp in ancient China. They were commented by Charles Darwin as “Passing over an almost infinite diversity of color, we meet with the most extraordinary modifications of structure”. Extremely diverse features and long history of domestication and breeding make goldfish an excellent genetic model system for fish physiology and evolution.
Achievements
Goldfish genome
Joint analysis of PacBio and Illumina pair-end sequencing data yields an initial 1.657 G draft assembly (Contig N50=474 Kb). Bionano optical map was generated and corrected the assembly into 1.73 Gb in size (Estimated genome size: 1.8 Gb). Hi-C based assembly further improved scaffold N50 from 606 Kb to 31.84 Mb and achieved 95.75% (1.65 Gb) oriented and ordered anchoring rate. The genome comprises 56,251 coding genes and 10,098 long non-coding transcript. Moreover, 38 potential centromeric regions were predicted out of 50 chromosomes.
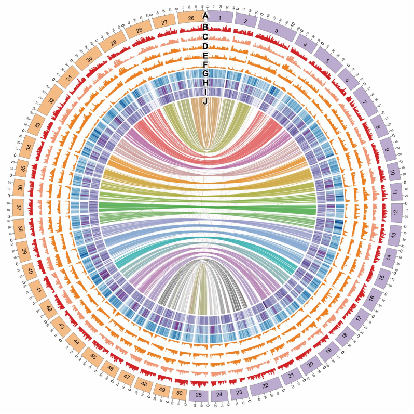
Fig.1 Gold fish Genome
Two clear sets of subgenomes were identified in the 50 goldfish chromosomes that resulted from an ancient hybridization event. The set of chromosomes with higher proportion of reads aligned between goldfish and Barbinae were defined as subgenome A (ChrA01~A25), i.e. subgenome common to Barbinae, and the remainings as subgenome B (ChrB01~B25).
Domestication and selective sweeps
A total of 16 wild type crucian carps and 185 representative goldfish variants were resequence with an average sequencing depth of approximately 12.5X, generating 4.3 terabases of data. Phylogenetic reconstruction and PCA analysis confirmed a closer relation between common goldfish and crucian carp than other goldfish, which latter divided into two lineages.
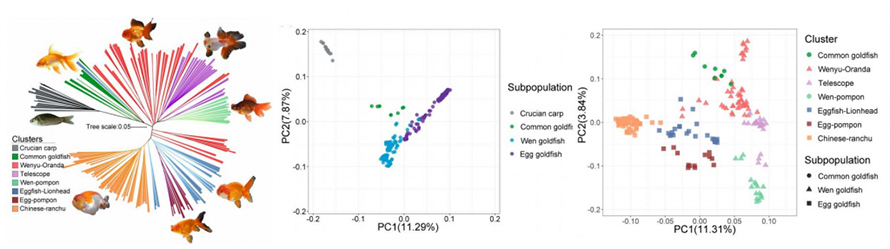
LD decay analysis on above four subpopulations supported the existence of a population genetic bottleneck during the domestication and strong artificial selection in goldfish. Increasing genetic diversity (π) from crucian carp to common goldfish further to Wen goldfish and Egg goldfish indicated a significant accumulation of genetic variations during their domestication. 50 selective sweep genomic regions covering 25.2 Mb and 946 genes were identified from representative data (33 goldfish and 16 crucian crap). Expanding analysis to 201 individuals, 393 genes indicated regions of completed selective sweep. These genes were found of low-diversity, which are likely contributed to the phenotypes associated with major domestication traits in goldfish.
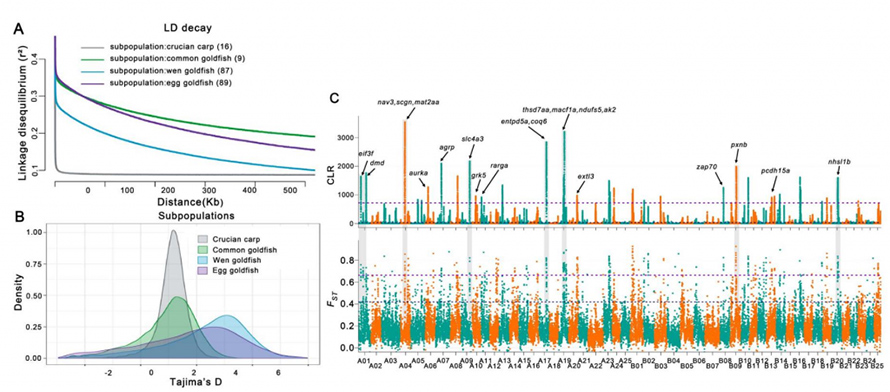
Fig.3 Genome-wide domestication-associated analysis
GWAS on domesticated goldfish
Dorsal fin is a key feature distinguishes Wen goldfish from Egg goldfish. GWAS of dorsal fin on 96 Wen goldfish and 87 Egg goldfish revealed 378 candidate genes spread across 13 chromosomes and an uneven distribution of these genes between subgenomes was observed. Functional analysis on these candidate genes highlighted biological processes including “Cell surface receptor signaling”, “Transmembrane transportation”, “Skeletal system development”, etc.
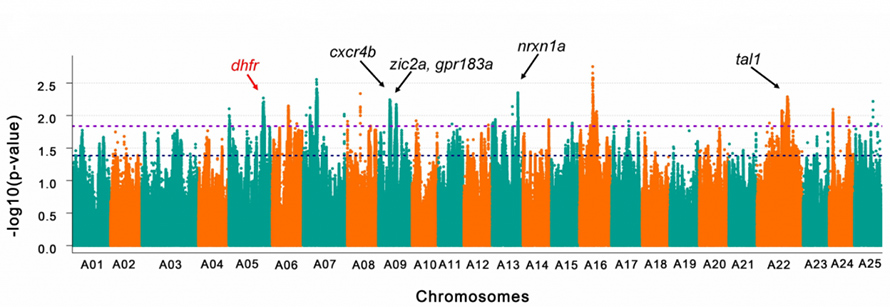
Fig.4 GWAS of dorsal fin on domesticated goldfish
In GWAS of transparent scale-related traits, a single strong association peak was detected. A gene encoding a tyrosine-protein kinase receptor was identified in one of candidate regions.
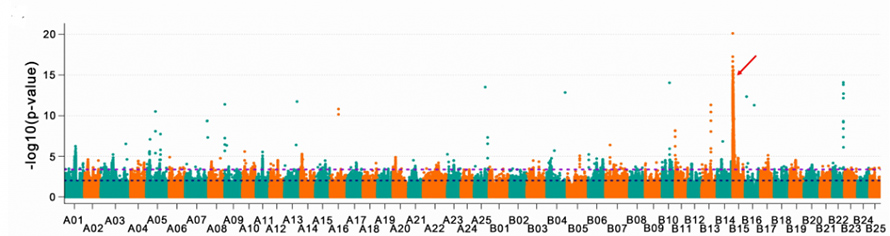
Fig.5 GWAS of transparent scale-related traits
Reference
Chen D et al. The evolutionary origin and domestication history of goldfish (Carassius auratus). PNAS (2020)
News aims at sharing the latest successful cases with Biomarker Technologies, capturing novel scientific achievements as well as prominent techniques applied during the study.
Post time: Jan-04-2022

