T2T GENOME ASSEMBLY, GAP FREE GENOME
1stMitundu iwiri ya Rice1
Mutu: Msonkhano ndi Kutsimikizika kwa Ma Genomes Awiri Opanda Gap a Xian/indica Rice Akuwulula Kuzindikira mu Zomangamanga za Plant Centromere
Doi:https://doi.org/10.1101/2020.12.24.424073
Yolembedwa Nthawi: Januware 01, 2021.
Institute: Huazhong Agricultural University, China
Zipangizo
O. sativa xian/indicamitundu ya mpunga 'Zhenshan 97 (ZS97)' ndi 'Minghui 63 (MH63)
Njira yotsatirira
NGS imawerenga + HiFi imawerenga + CLR imawerenga + BioNano + Hi-C
Zambiri:
ZS97: 8.34 Gb (~ 23x) HiFi imawerenga + 48.39 Gb (~131x ) CLR imawerenga + 25 Gb(~69x) NGS + 2 Maselo a BioNano Irys
MH63: 37.88 Gb (~103x) HiFi imawerenga + 48.97 Gb (~132x) CLR imawerenga + 28 Gb(~76x) NGS + 2 ma cell a BioNano Irys
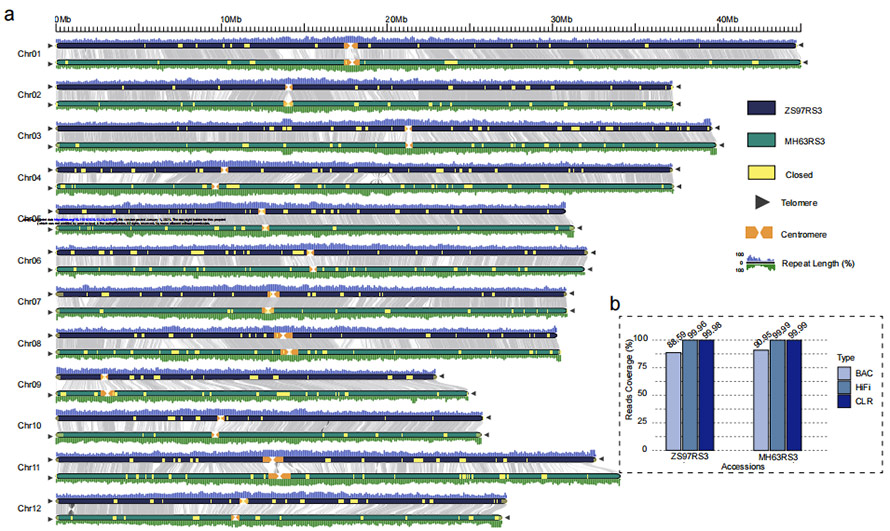
Chithunzi 1 Ma genome awiri opanda malire a mpunga (MH63 ndi ZS97)
2ndBanana Genome2
Kamutu: Telomere-to-telomere gapless chromosome ya nthochi pogwiritsa ntchito nanopore sequencing
Doi:https://doi.org/10.1101/2021.04.16.440017
Yolembedwa Nthawi: Epulo 17, 2021.
Institute: Université Paris-Saclay, France
Zipangizo
Haploid iwiriMusa acuminatasppmatenda a malaccensis(DH-Pahang)
Njira zotsatirira ndi deta:
HiSeq2500 PE250 mode + MinION/ PromethION (93Gb, ~200X )+ Mapu opangira (DLE-1+BspQ1)
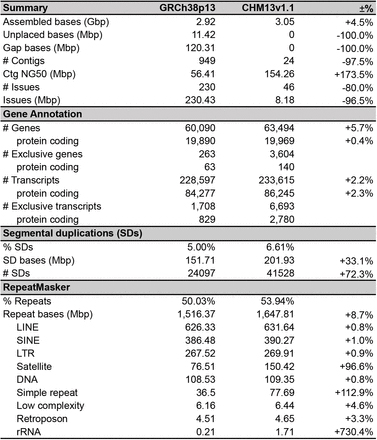
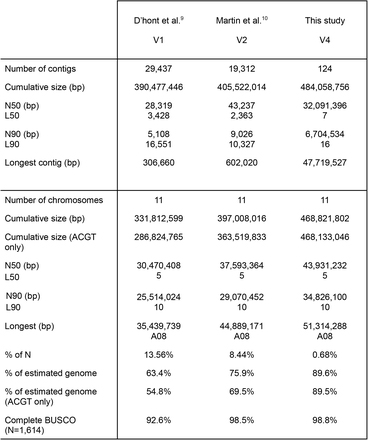
Table 1 Kuyerekeza kwa Musa acuminata (DH-Pahang) genome assemblies
Chithunzi 2 Musa genomes kufananitsa zomangamanga
3rdPhaeodactylum tricornutum genome3
Mutu: Msonkhano wa genome wa Telomere-to-telomere waP
heodactylum tricornutum
Doi:https://doi.org/10.1101/2021.05.04.442596
Yolembedwa Nthawi: Meyi 04, 2021
Institute: Western University, Canada
Zipangizo
Phaeodactylum tricornutum(Culture Collection of Algae and Protozoa CCAP 1055/1)
Njira zotsatirira ndi deta:
1 Oxford Nanopore minion flow cell + 2 × 75 yophatikizika-mapeto apakati pazotulutsa NextSeq 550 run
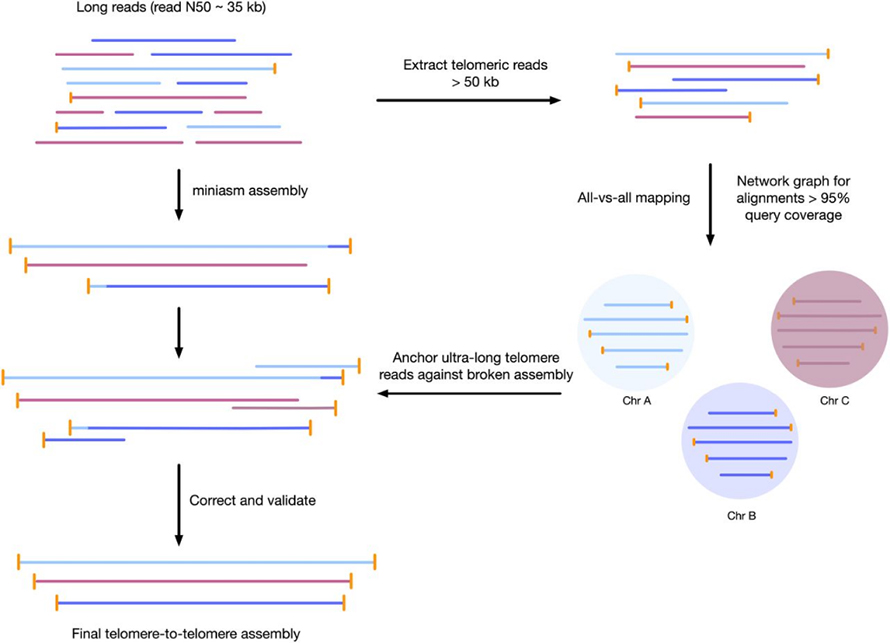
Chithunzi 3 Kayendetsedwe ka ntchito ka telomere-to-telomere genome assembly
4thMunthu CHM13 genome4
Kamutu: Mndandanda wathunthu wa jini la munthu
Doi:https://doi.org/10.1101/2021.05.26.445798
Yolembedwa Nthawi: Meyi 27, 2021
Institute: National Institutes of Health (NIH), USA
Zida: cell line CHM13
Njira zotsatirira ndi deta:
30 × PacBio circular consensus sequencing (HiFi), 120 × Oxford Nanopore Ultra-long read sequencing , 100 × Illumina PCR-Free sequencing (ILMN) , 70 × Illumina / Arima Genomics Hi-C (Hi-C), BioNano ndi Strand-seq
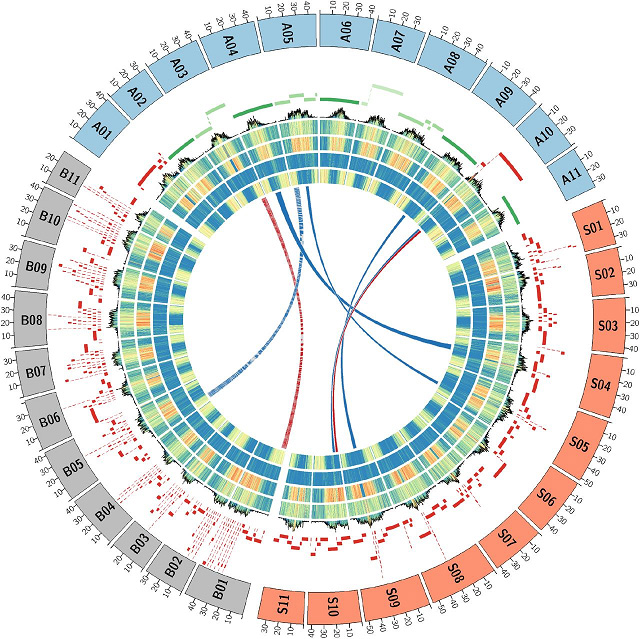
Gulu 2 Kuyerekeza kwa GRCh38 ndi T2T-CHM13 magulu amtundu wa anthu
Buku
1.Sergey Nurk et al.Mndandanda wathunthu wa jini la munthu.bioRxiv 2021.05.26.445798;doi:https://doi.org/10.1101/2021.05.26.445798
2.Caroline Belser et al.Telomere-to-telomere gapless chromosomes ya nthochi pogwiritsa ntchito nanopore sequencing.bioRxiv 2021.04.16.440017;doi:https://doi.org/10.1101/2021.04.16.440017
3.Daniel J. Giguere et al.Msonkhano wa telomere-to-telomere genome wa Phaeodactylum tricornutum.bioRxiv 2021.05.04.442596;doi:https://doi.org/10.1101/2021.05.04.442596
4.Jia-Ming Song et al.Msonkhano ndi Kutsimikizika kwa Ma Genome Awiri Opanda Gap a Reference Genomes a Xian/indica Rice Akuwulula Kuzindikira mu Zomangamanga za Plant Centromere.bioRxiv 2020.12.24.424073;doi:https://doi.org/10.1101/2020.12.24.424073
Nthawi yotumiza: Jan-06-2022

