T2T GENOME ASSEMBLY, GAP FREE GENOME
1st Two Rice Genomes1
Title: Assembly and Validation of Two Gap-free Reference Genomes for Xian/indica Rice Reveals Insights into Plant Centromere Architecture
Doi: https://doi.org/10.1101/2020.12.24.424073
Posted Time: January 01, 2021.
Institute: Huazhong Agricultural University, China
Materials
O. sativa xian/indica rice varieties ‘Zhenshan 97 (ZS97)’ and ‘Minghui 63 (MH63)
Sequencing strategy
NGS reads + HiFi reads + CLR reads + BioNano + Hi-C
Data:
ZS97: 8.34 Gb(~23x)HiFi reads + 48.39 Gb (~131x ) CLR reads + 25 Gb(~69x) NGS + 2 BioNano Irys cells
MH63: 37.88 Gb (~103x) HiFi reads + 48.97 Gb (~132x) CLR reads + 28 Gb(~76x) NGS + 2 BioNano Irys cells
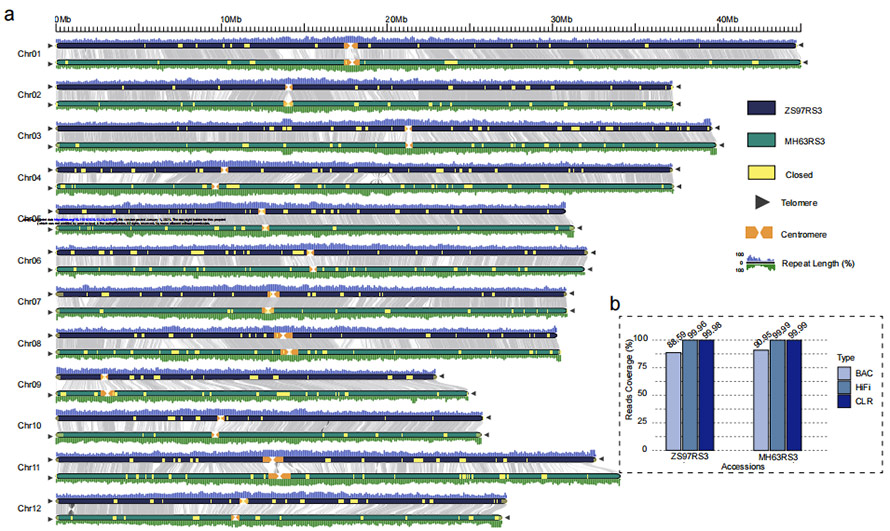
Figure 1 Two gap-free genomes of rice (MH63 and ZS97)
2nd Banana Genome2
Title: Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing
Doi: https://doi.org/10.1101/2021.04.16.440017
Posted Time: April 17, 2021.
Institute: Université Paris-Saclay, France
Materials
Double haploid Musa acuminata spp malaccensis (DH-Pahang)
Sequencing strategy and data:
HiSeq2500 PE250 mode + MinION/ PromethION (93Gb,~200X )+ Optical map (DLE-1+BspQ1)
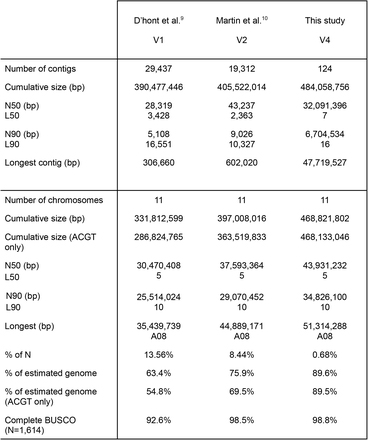
Table 1 Comparison of Musa acuminata (DH-Pahang) genome assemblies
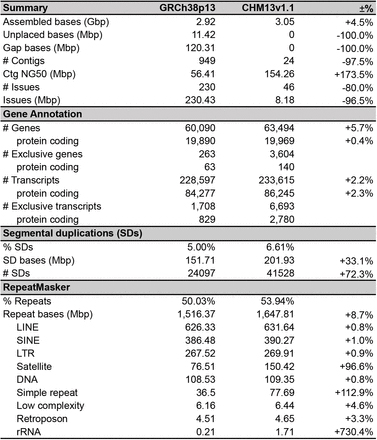
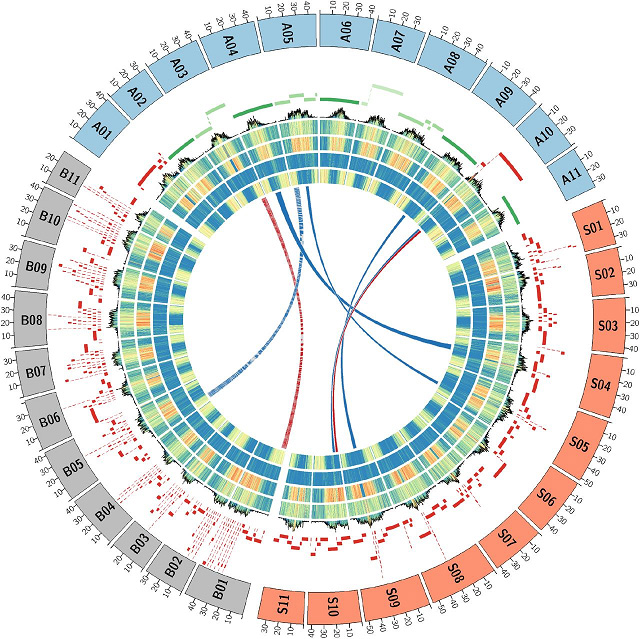
Figure 2 Musa genomes architecture comparison
3rd Phaeodactylum tricornutum genome3
Title: Telomere-to-telomere genome assembly of P
haeodactylum tricornutum
Doi: https://doi.org/10.1101/2021.05.04.442596
Posted Time: May 04, 2021
Institute: Western University, Canada
Materials
Phaeodactylum tricornutum (Culture Collection of Algae and Protozoa CCAP 1055/1)
Sequencing strategy and data:
1 Oxford Nanopore minION flow cell + a 2×75 paired-end mid-output NextSeq 550 run
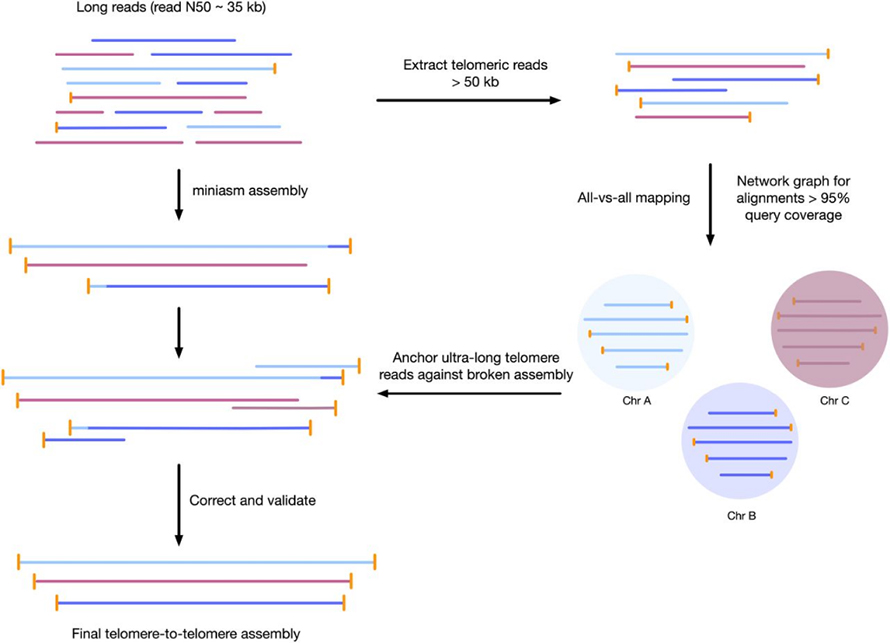
Figure 3 Workflow for telomere-to-telomere genome assembly
4th Human CHM13 genome4
Title: The complete sequence of a human genome
Doi: https://doi.org/10.1101/2021.05.26.445798
Posted Time: May 27, 2021
Institute: National Institutes of Health (NIH), USA
Materials: cell line CHM13
Sequencing strategy and data:
30× PacBio circular consensus sequencing (HiFi) , 120× Oxford Nanopore ultra-long read sequencing , 100× Illumina PCR-Free sequencing (ILMN) , 70× Illumina / Arima Genomics Hi-C (Hi-C), BioNano optical maps, and Strand-seq
Table 2 Comparison of GRCh38 and T2T-CHM13 human genome assemblies
Reference
1.Sergey Nurk et al. The complete sequence of a human genome. bioRxiv 2021.05.26.445798; doi: https://doi.org/10.1101/2021.05.26.445798
2.Caroline Belser et al. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. bioRxiv 2021.04.16.440017; doi: https://doi.org/10.1101/2021.04.16.440017
3.Daniel J. Giguere et al. Telomere-to-telomere genome assembly of Phaeodactylum tricornutum. bioRxiv 2021.05.04.442596; doi: https://doi.org/10.1101/2021.05.04.442596
4.Jia-Ming Song et al. Assembly and Validation of Two Gap-free Reference Genomes for Xian/indica Rice Reveals Insights into Plant Centromere Architecture. bioRxiv 2020.12.24.424073; doi: https://doi.org/10.1101/2020.12.24.424073
Post time: Jan-06-2022

