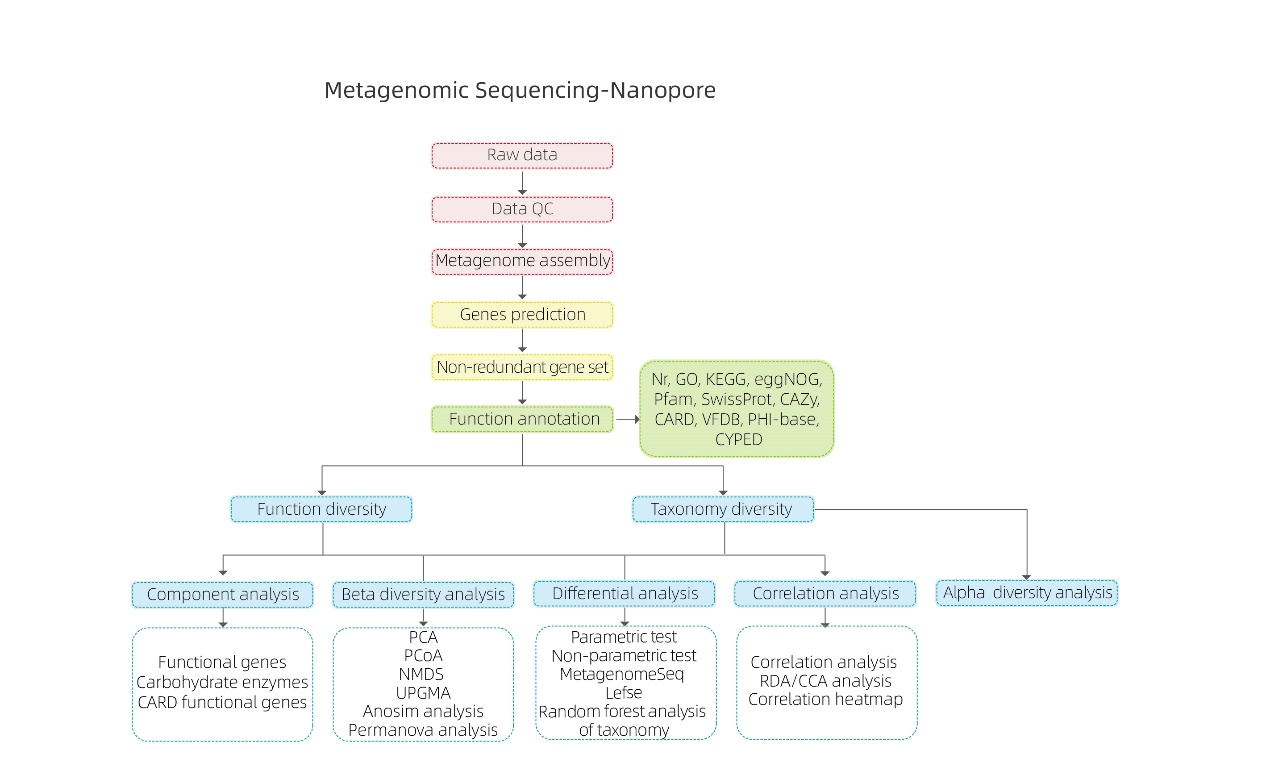
Metagenomic Sequencing-Nanopore
Servitium Commoda
High-qualitas congregationis Enhancing accurate speciei identificatio et funcational gene praedictio
Clausa bacterial genome solitudo
Potentior et certa applicatio in diversa area, eg deprehensio microorganismi pathogenici vel resistentiae antibioticae genesis cognatae.
Metagenoma comparativum analysis
Service Specifications
| Platform | Sequencing | Commendatur data | Tunc turnaround |
| Nanopore | ONT | 6 G/10 G | LXV diebus ferialibus |
Bioinformatics analyses
Rudis notitia qualis imperium
Metagenome conventus
Non-abundans gene set et annotation
Species diversitatis analysis
Munus geneticae diversitatis analysis
inter coetus analysis
● Analysis Association contra factores experimentales
Sample Requisita et Delivery
Sample requisita et traditio
Sample Requisita:
ForDNA excerpta:
| Sample Type | Moles | Coniunctis | Puritas |
| DNA excerpta | 1-1.5 μg | 20 ng/µl | OD260/280= 1.6-2.5 |
Ad exempla environmental:
| Sample type | Commendatur sampling procedure |
| Solum | Sampling amount: approx.5 g;Manens autem substantia arida debet a superficie removeri;Tere magna frusta et per 2 mm sparguntur;Aliquot specimina in EP-tube vel cyrotube sterili ad reservationem. |
| feces | Sampling amount: approx.5 g;Collecta et aliquot exempla in EP-tube vel cryotube sterili ad reservationem. |
| Intestina contenta | Exempla necesse est sub conditione aseptic discursum.Lava collecta cum PBS;Centrifuge PBS et praecipitantem in EP-tubes collige. |
| Sludge | Sampling amount: approx.5 g;Collecta et aliquot pituitae specimen in sterili ep-tube vel cryotube ad reservationem |
| Waterbody | Specimen pro limitata copia microbialium, ut sonum aquae, putei aquae, etc., collige saltem 1 L aquam et per 0.22 µm colum transi, ut microbialem in membrana locupletet.Membranam in tubo sterili repone. |
| Cutis | Superficiem cutem diligenter corradunt cum swab bombicis sterilibus vel chirurgicis mucrone et in tubo sterili pone. |
Sample Delivery commendatur
Exempla in nitrogenio liquido per 3—4 horas durata et in liquido nitrogenio vel -80 gradu ad longi temporis reservationem reponunt.Sample naviculas cum sicco-glacie exquiritur.
Service Opus O

Sample traditio

Library construction

Sequencing

Analysis

Post-venditionis officia
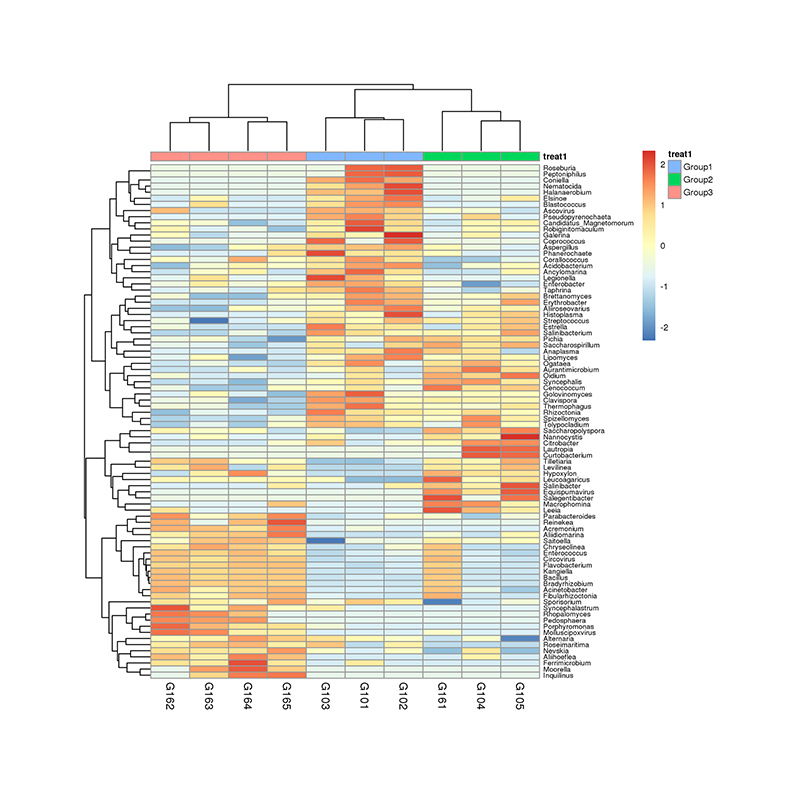
1.Heatmap: species pampineis pampineis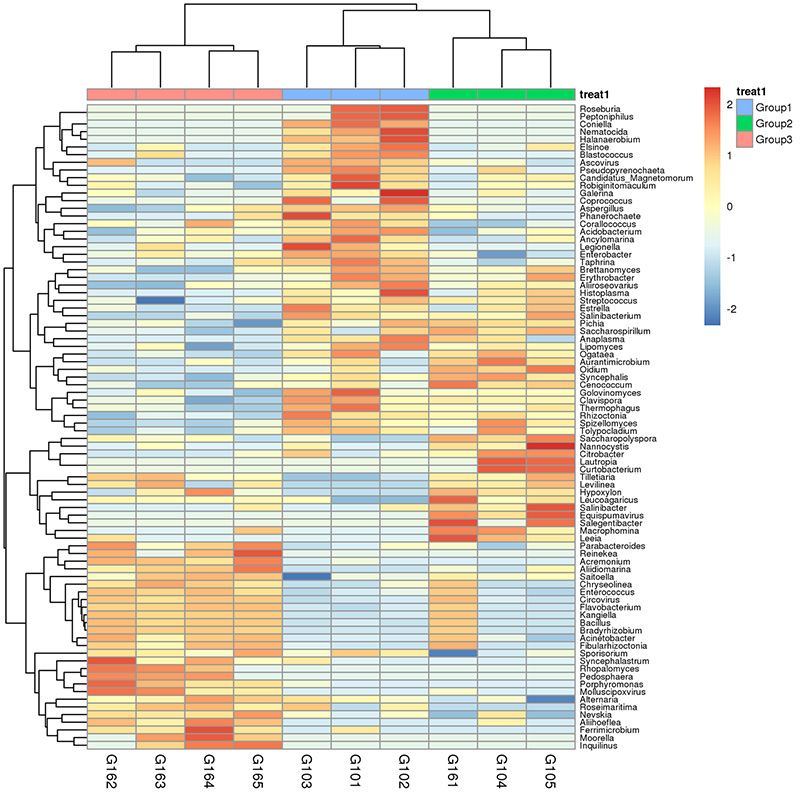 2.Functional genes annotatae ad KEGG metabolicae meatus
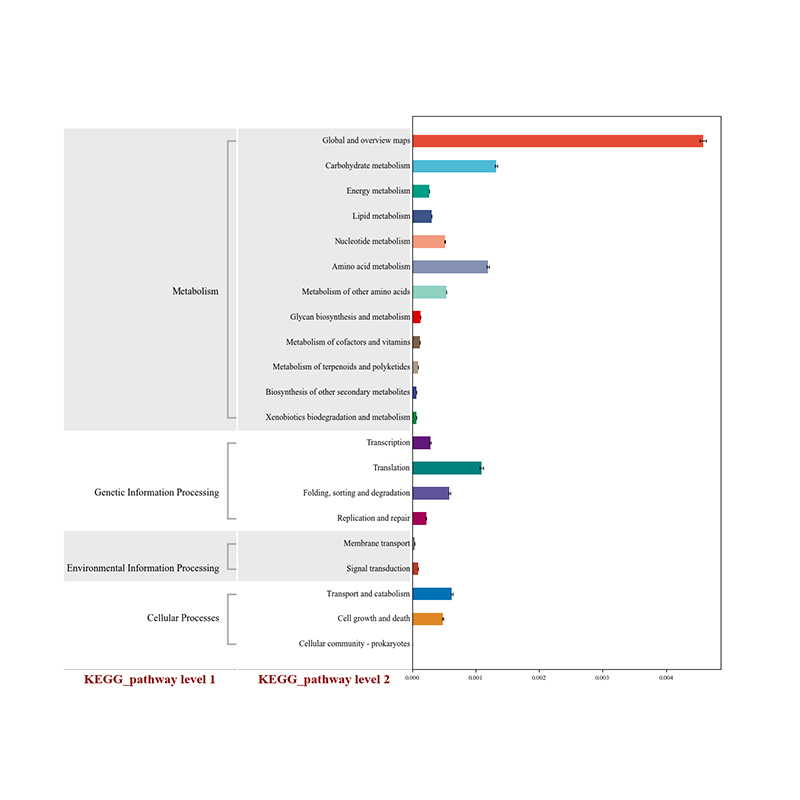
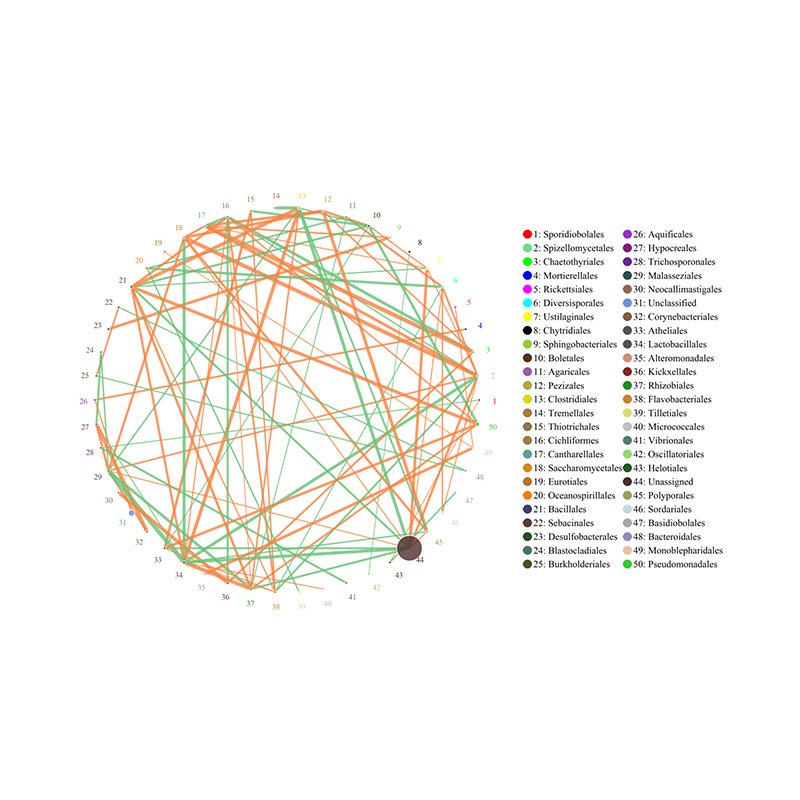
2.Functional genes annotatae ad KEGG metabolicae meatus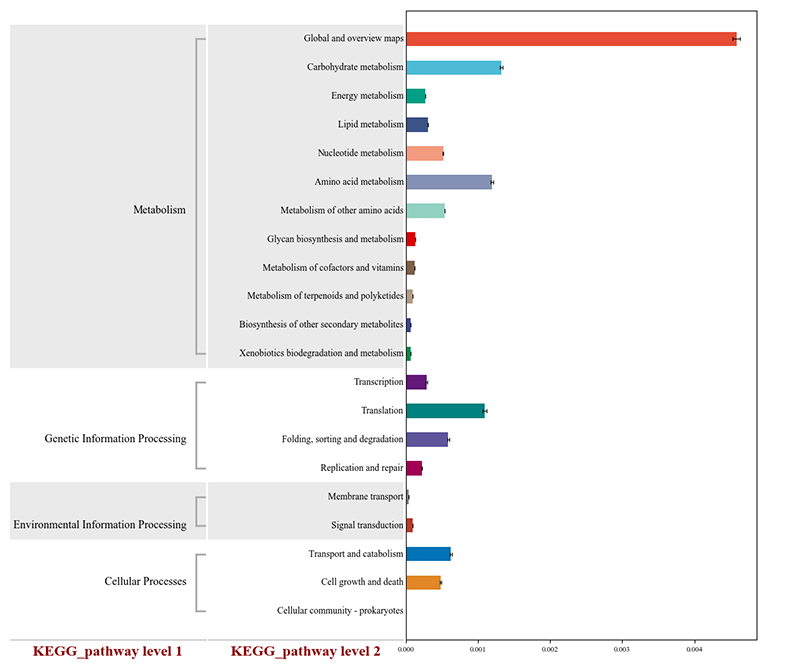 3.Species ratione network
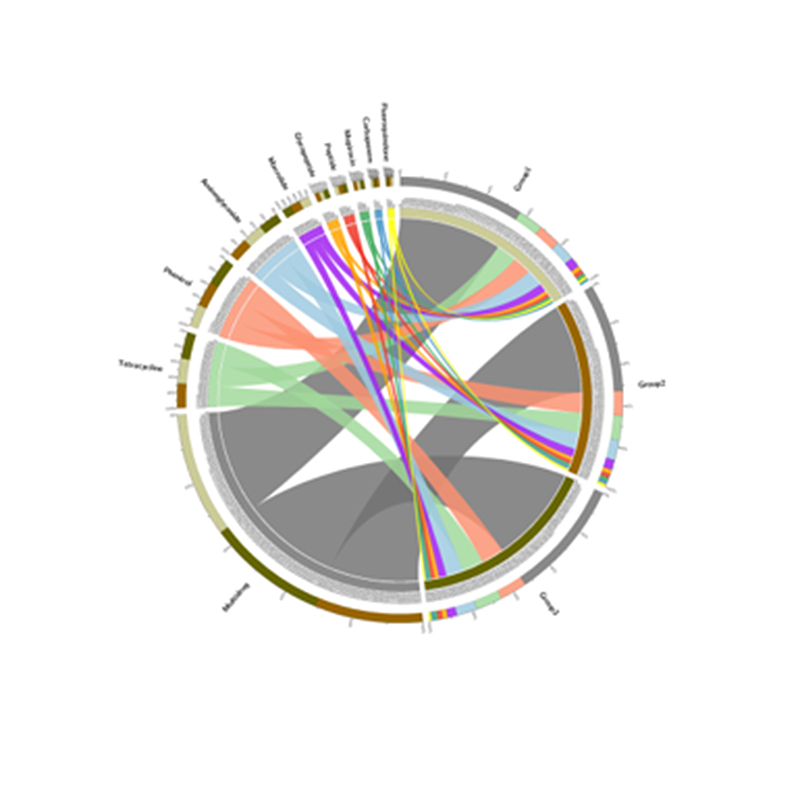
3.Species ratione network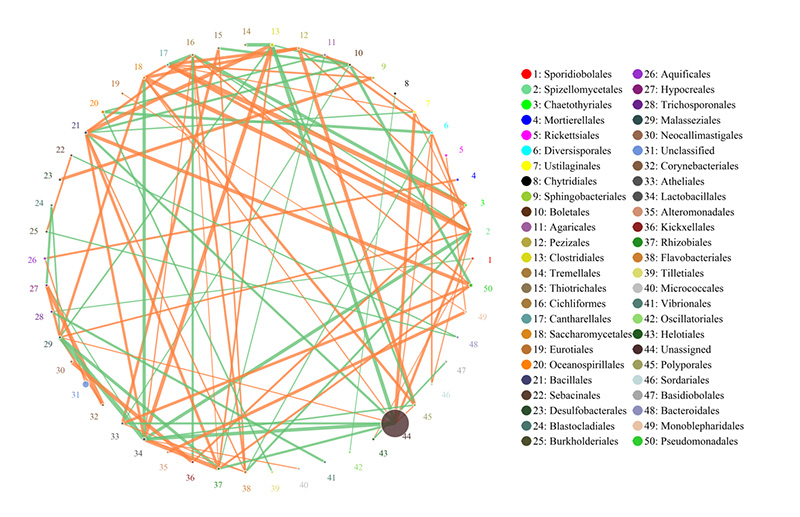 4.Circos card resistentiae antibioticae genes
4.Circos card resistentiae antibioticae genes
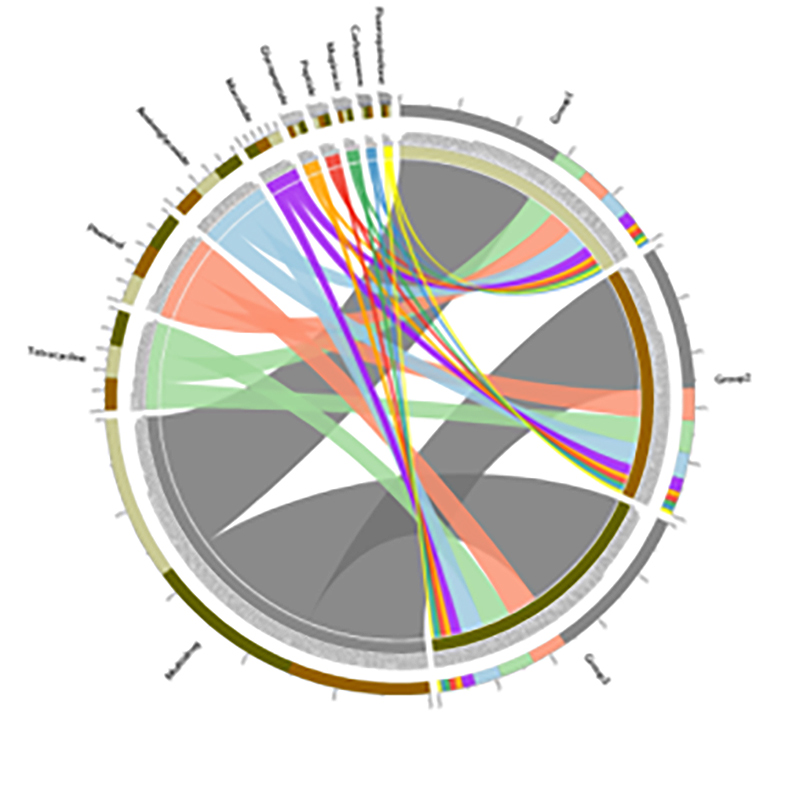
BMK Case
Nanopore metagenomica celeris clinicis diagnosis bacterial contagione respiratorii inferioris dat
Published:Natura Biotechnologia, 2019
Technical volutpat
Sequentia: Nanopore MinION
Orci metagenomici bioinformatici: Hostia DNA deperditio, WIMP et ARMA analysis
Celeri deprehensio: VI horas
Summus sensus: 96.6%
Eventus key
Anno 2006, infectiones respiratoriae inferioris (LR) III decies centena milia mortis humanae globaliter effecit.Methodus typica deprehendendi LR1 pathogen est cultura, quae pauper sensus, longi vicis et moderatio caret in therapia antibiotica prima.Celeris et accurata diagnosis microbialis diu urgente necessitate fuit.Dr. Justinus ex Universitate Orientalis Angliae et sociis suis a Nanopore fundato methodum metagenomicam ad pathogen deprehendendi feliciter explicavit.Secundum eorum workflow, 99,99% exercitus DNA levari potest.Deprehensio in genesis pathogenis et antibioticis repugnantibus in 6 horis finiri potest.
Reference
Charalampous, T. , Kay, GL , Richardson, H. , Aydin, A. , & O'Grady, J. .(2019).Nanopore metagenomica celeris clinicis diagnosis bacterial contagione respiratorii inferioris efficit.Naturae Biotechnologiae, 37 (7), 1.