
Hi-C yakavakirwa Genome Assembly
Basa Rinobatsira
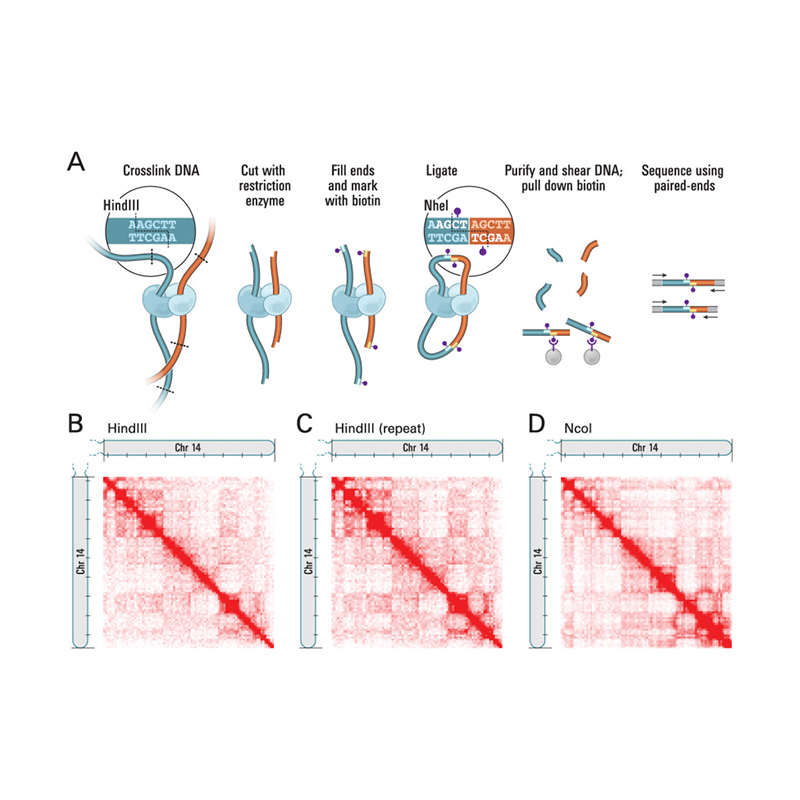
Mhedziso yeHi-C
(Lieberman-Aiden Et al.,Sayenzi, 2009)
● Hapana chikonzero mukugadzira genetic population ye contig anchoring;
● Higher marker density inotungamira kune yakakwira contigs anchoring ratio iri pamusoro pe90%;
● Inogonesa kuongororwa uye kugadzirisa pamagungano ejenome aripo;
● Kupfupika-kutenderera-nguva nepamusoro-soro mukusangana kwegenome;
● Ruzivo rwakawanda nemaraibhurari eHi-C anopfuura 1000 akavakirwa marudzi anopfuura mazana mashanu;
● Mhosva dzinopfuura zana dzakabudirira dzine accumulative published impact factor inopfuura 760;
● Hi-C based genome assembly yepolyploid genome, 100% anchoring rate yakawanikwa mupurojekiti yapfuura;
● In-house patents uye software copyrights yeHi-C kuedza uye kuongorora data;
● Kuzvigadzira-kuzvigadzira kwekuona data tuning software, inogonesa manual block kufamba, kudzosera kumashure, kudzoreredza uye kuitazve.
Sevhisi Zvinotsanangurwa
|
Raibhurari Type
|
Platform | Read Length | Kurudzira Strategy |
| Hi-C | Illumina NovaSeq | PE150 | ≥ 100X |
Bioinformatics inoongorora
● Raw data quality control
● Hi-C raibhurari yemhando yekudzora
● Hi-C based genome assembly
● Kuongororwa kwepashure pegungano
Sample Zvinodiwa uye Delivery
Sample Zvinodiwa:
| Mhuka | Fungus | Zvirimwa
|
| Matishu akaomeswa nechando: 1-2g paraibhurari Masero: 1x 10^7 maseru paraibhurari | Matishu akaomeswa nechando: 1g paraibhurari | Matishu akaomeswa nechando: 1-2g paraibhurari
|
| *Tinokurudzira kutumira angangoita 2 aliquots (1 g yega) yeHi-C kuyedza. | ||
Yakakurudzirwa Sample Delivery
Container: 2 ml centrifuge chubhu (Tin foil haina kukurudzirwa)
Kune akawanda emasampuli, isu tinokurudzira kuti usachengetedze mu ethanol.
Sample labeling: Samples dzinofanirwa kunyorwa zvakajeka uye dzakafanana neyakatumirwa ruzivo ruzivo fomu.
Shipment: Dry-ice: Samples dzinofanirwa kurongedzerwa mumabhegi kutanga uye kuvigwa mune yakaoma-aizi.
Kuyerera Kwebasa Rebasa

Kuedza kugadzira

Muenzaniso wekutumira

Kutorwa kweDNA

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
*Demo mhedzisiro inoratidzwa pano yese kubva kugenomes yakaburitswa neBiomarker Technologies
1.Hi-C kudyidzana kupisa mepu yeCamptotheca acuminatagenome.Sezvinoratidzwa pamepu, kuwanda kwekudyidzana kwakabatana zvisina kunaka nemutsara chinhambwe, icho chinoratidza gungano rakanyanya-chaiyo chromosome-level.(Anchoring ratio: 96.03%)
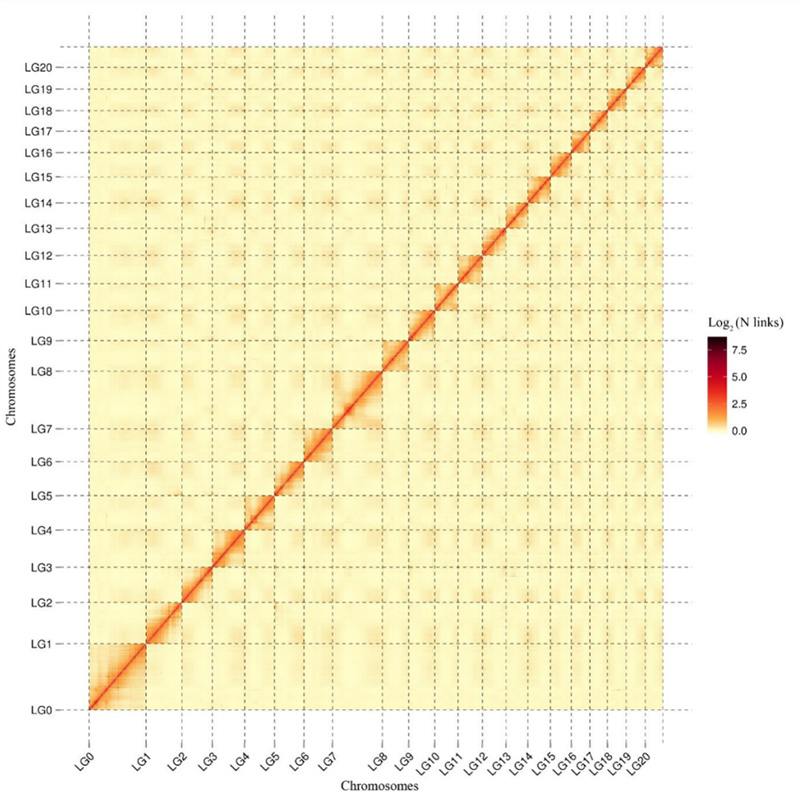
Kang M et al.,Nature Communications, 2021
2.Hi-C yakafambisa kusimbiswa kweinversions pakatiGossypium hirsutumL. TM-1 A06 uyeG. arboreumChr06
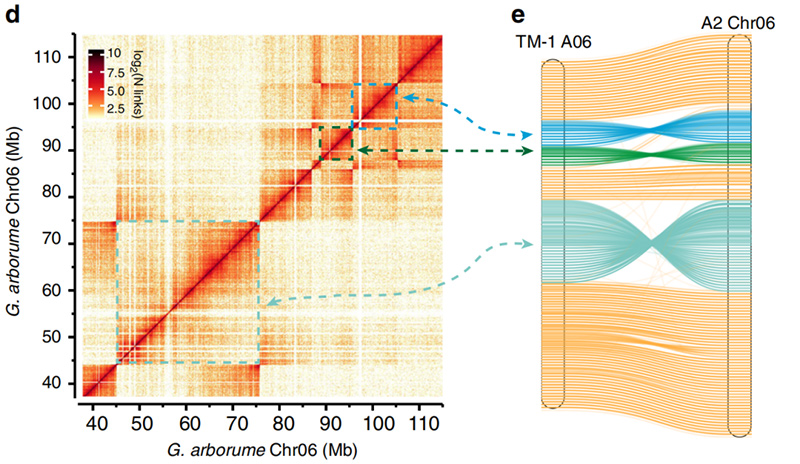
Yang Z et al.,Kukurukurirana Nezvakasikwa, 2019
3.Assembly and biallelic differentiation yemufarinya genome SC205.Hi-C heatmap inoratidzwa zvakajeka kupatsanurwa mumakromosomes ane homologous.
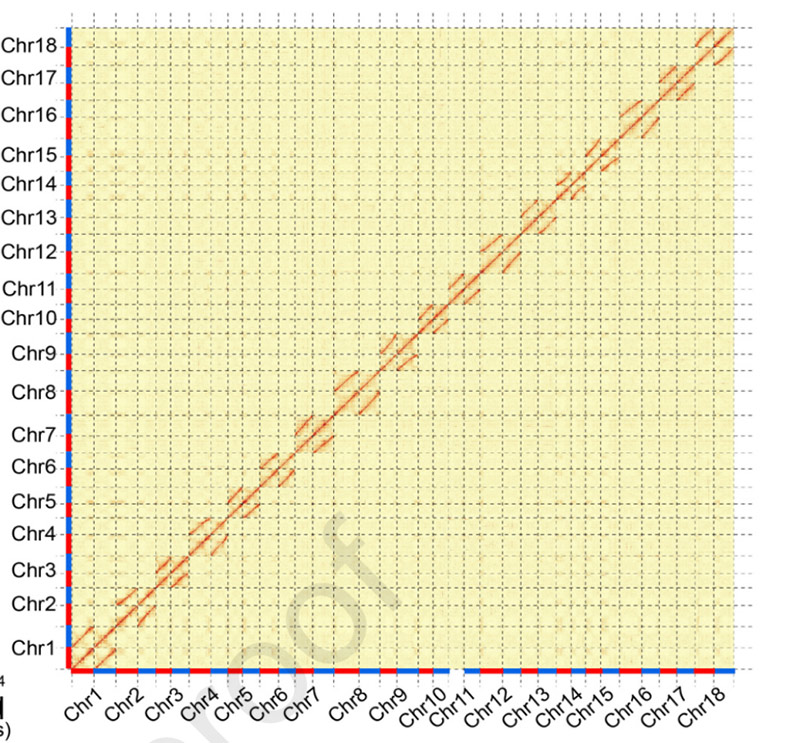
Hu W et al.,Molecular Plant, 2021
4.Hi-C heatmap pamhando mbiri dzeFicus genome assembly:F.microcarpa(anchoring ratio: 99.3%) uyeF.hispida (anchoring ratio: 99.7%)
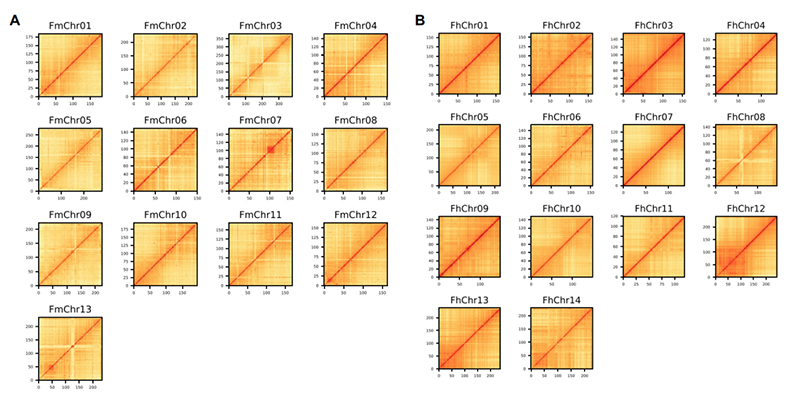
Zhang X et al.,Sero, 2020
Nyaya yeBMK
Genomes Emuti weBanyan uye Pollinator Wasp Inopa Manzwisisiro MuFig-wasp Coevolution
Rakatsikiswa: Sero, 2020
Sequencing strategy:
F. microcarpa genome: Approx.84 X PacBio RSII (36.87 Gb) + Hi-C (44 Gb)
F. hispidagenome: Approx.97 X PacBio RSII (36.12 Gb) + Hi-C (60 Gb)
Ekudzidzisana verticillatagenome: Approx.170 X PacBio RSII (65 Gb)
Mibairo yakakosha
1.Two banyan tree genomes uye imwe pollinator wasp genome zvakavakwa pachishandiswa PacBio sequencing, Hi-C uye yekubatanidza mepu.
(1)F. microcarpagenome: Gungano re426 Mb (97.7% yeinofungidzirwa genome saizi) yakavambwa ine contig N50 ye908 Kb, BUSCO mamakisi e95.6%.Pakazara 423 Mb kutevedzana kwakamiswa kune gumi nematatu machromosomes neHi-C.Genome annotation yakaburitsa 29,416 protein-coding genes.
(2)F. Hispidagenome: Gungano re360 Mb (97.3% yeinofungidzirwa genome saizi) yakapihwa necontig N50 ye492 Kb uye BUSCO mamakisi e97.4%.Huwandu hwe359 Mb kutevedzana kwakadzikwa pamakromosomes gumi nemana neHi-C uye dzakanyanya kufanana nemepu yekubatanidza yakakwira-density.
(3)Ekudzidzisana verticillatagenome: Gungano re387 Mb (Inofungidzirwa saizi yegenome: 382 Mb) yakavambwa ine contig N50 ye3.1 Mb uye BUSCO mamakisi e97.7%.
2.Comparative genomics ongororo yakaratidza huwandu hukuru hwemagadzirirwo akasiyana pakati pezviviriFicusgenomes, iyo yakapa yakakosha genetic resource ye adaptive evolution zvidzidzo.Ichi chidzidzo, kekutanga, chakapa nzwisiso muFig-wasp coevolution pagenomic-level.
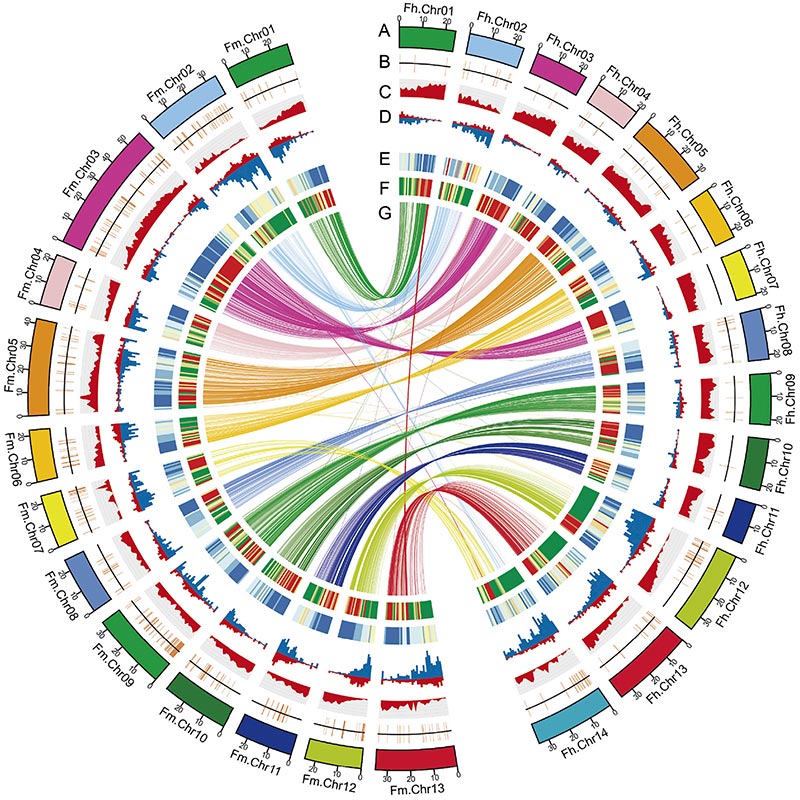 Circos dhayagiramu pane genomic maficha maviriFicusgenomes, kusanganisira machromosomes, segmental duplications (SDs), transposons(LTR, TEs, DNA TEs), gene expression uye synteny. | 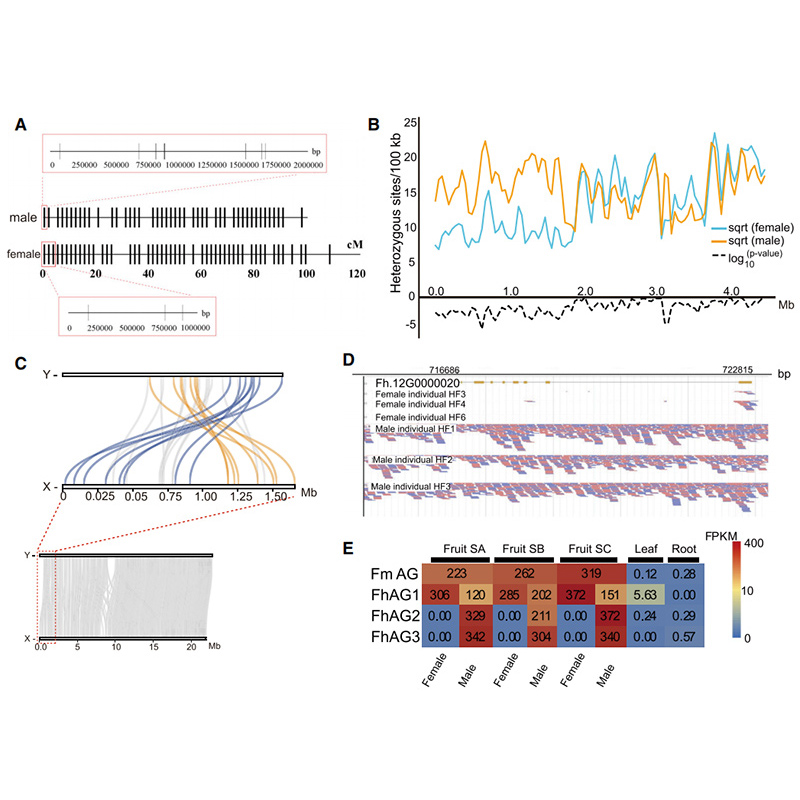 Kuzivikanwa kweiyo Y chromosome uye bonde kutsunga mumiriri gene |
Zhang, X, nevamwe."Genomes yeBanyan Tree uye Pollinator Wasp Inopa Manzwisisiro muFig-Wasp Coevolution."Sero 183.4(2020).














