Metagenomic Sequencing -NGS
Basa Rinobatsira
● Kuzviparadzanisa nevamwe uye kurima-pasina kune microbial nharaunda profiling
● Kugadziriswa kwepamusoro pakuona mhando dzakaderera-dzakawanda mumasampuli ezvakatipoteredza
● Pfungwa ye "meta-" inobatanidza zvinhu zvose zvehupenyu pahutano hwehutano, huwandu hwemhando uye gene level, iyo inoratidza maonero ane simba ari pedyo nechokwadi.
● BMK inounganidza ruzivo rwakakura mumhando dzakasiyana-siyana dzemuenzaniso dzine zvinopfuura 10,000 samples dzakagadziriswa.
Sevhisi Zvinotsanangurwa
| Platform | Sequencing | Yakakurudzirwa data | Nguva yekuchinja |
| Illumina NovaSeq Platform | PE150 | 6 G/10 G/20 G | 45 Mazuva ekushanda |
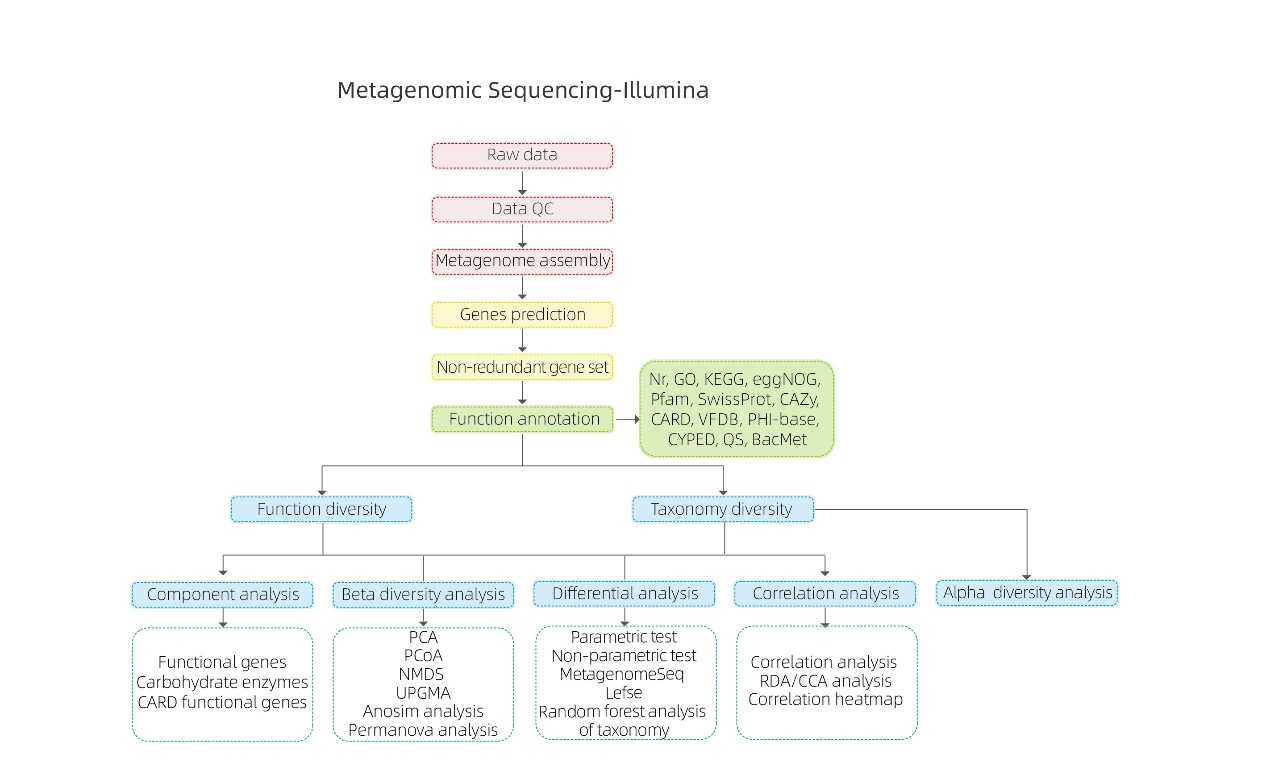
Bioinformatics inoongorora
● Raw data quality control
● Metagenome musangano
● Non-redundant gene set uye annotation
● Kuongororwa kwemhando dzakasiyana-siyana
● Genetic function diversity analysis
● Kuongororwa kweboka
● Kuongororwa kwesangano kunopesana nekuedza zvinhu
Sample Zvinodiwa uye Delivery
Sample Zvinodiwa:
ForDNA zvinyorwa:
| Muenzaniso Type | Mari | Concentration | Kuchena |
| DNA zvinyorwa | > 30 ng | > 1 ng/μl | OD260/280= 1.6-2.5 |
Zvezvakatipoteredza samples:
| Muenzaniso mhando | Inokurudzirwa sampling maitiro |
| Ivhu | Sampling huwandu: approx.5 g;Chinhu chakasara chakasvava chinoda kubviswa pamusoro;Geda zvidimbu zvakakura uye upfuure ne2 mm sefa;Aliquot samples mune sterile EP-chubhu kana cyrotube yekuchengetera. |
| Feces | Sampling huwandu: approx.5 g;Unganidza uye aliquot samples mune isinga bereki EP-tube kana cryotube yekuchengetera. |
| Intestinal contents | Samples dzinofanirwa kugadziriswa pasi peaseptic mamiriro.Geza matishu akaunganidzwa nePBS;Centrifuge iyo PBS uye tora iyo precipitant mu EP-machubhu. |
| Sludge | Sampling huwandu: approx.5 g;Unganidza uye aliquot sludge sampuli mune isina utachiona EP-tube kana cryotube yekuchengetera |
| Mvura | Yemuenzaniso ine huwandu hushoma hwehutachiona, senge mvura yemupombi, mvura yemugodhi, nezvimwewo, Unganidza ingangoita 1 L mvura uye upfuure nepakati pe0.22 μm sefa yekupfumisa hutachiona pa membrane.Chengetedza membrane muchubhu isina utachiona. |
| Ganda | Nyatsokweva pamusoro peganda nemucheka wedonje usina utachiona kana blade yekuvhiya woiisa muchubhu isina utachiona. |
Yakakurudzirwa Sample Delivery
Svitsa masampula munitrogen yemvura kwemaawa 3-4 uye chengetedza mune yemvura nitrogen kana -80 dhigirii kusvika kurebesa kwenguva refu.Sample shipping with dry-ice inodiwa.
Kuyerera Kwebasa Rebasa

Muenzaniso wekutumira

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
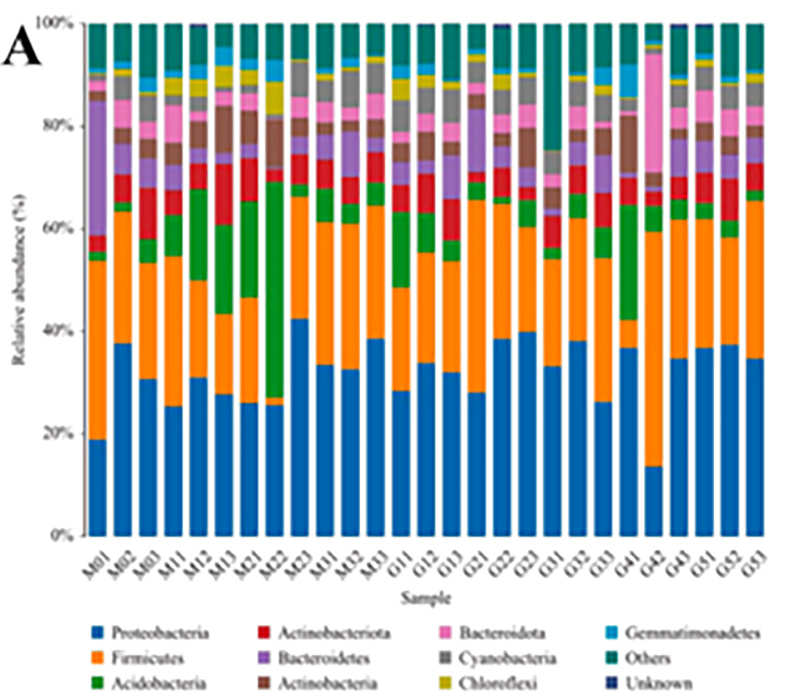
1.Histogram: Kugoverwa kwemarudzi
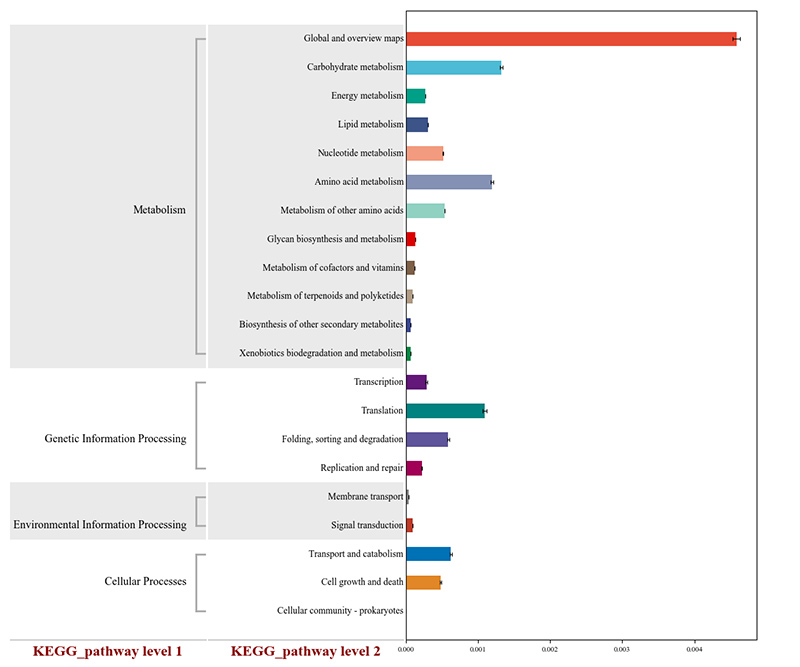
2.Magetsi anoshanda anotsanangurwa kuKEGG metabolic pathways
3.Heat mepu: Mabasa akasiyana zvichienderana nehuwandu hwemajini akawanda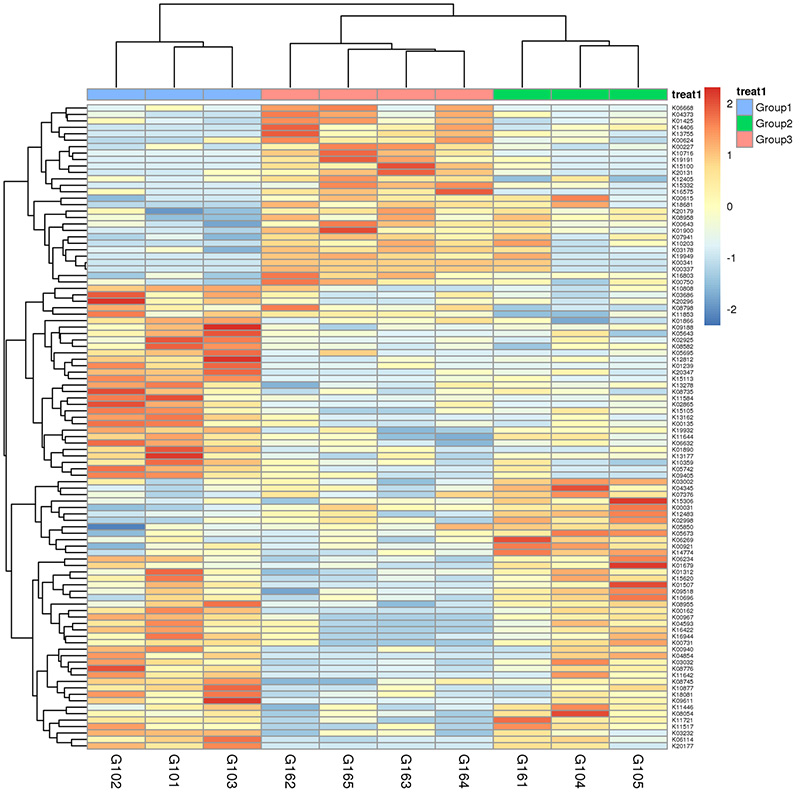 4.Circos yeCARD antibiotic resistance genes
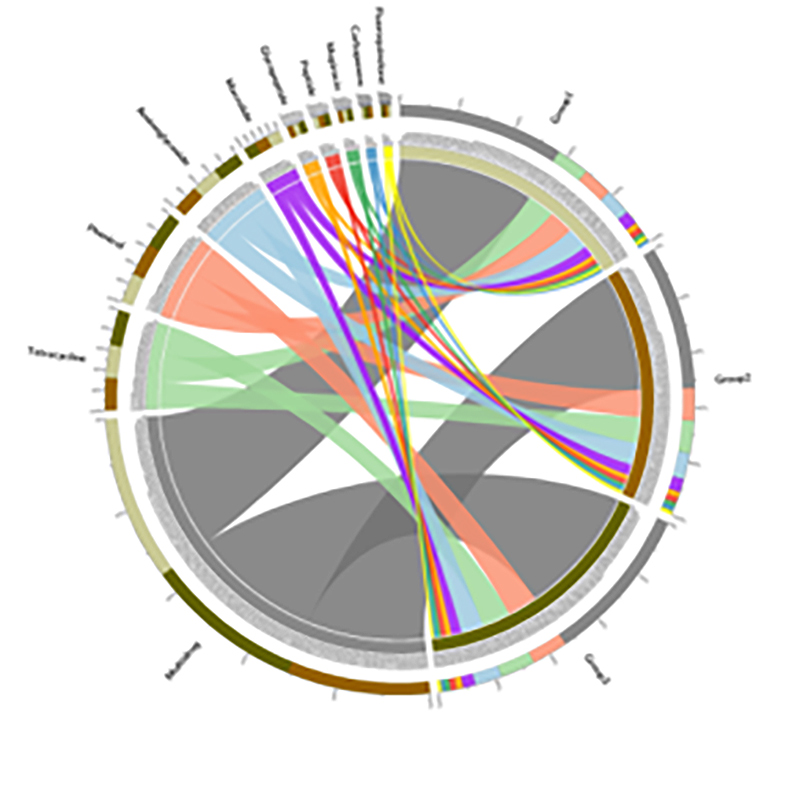
4.Circos yeCARD antibiotic resistance genes
Nyaya yeBMK
Kuwanda kwemajini ekudzivirira kwemishonga inorwisa mabhakitiriya uye hutachiona hwehutachiona pamwe nevhu-mangrove midzi inoenderera.
Rakatsikiswa:Chinyorwa cheHazardous Materials, 2021
Sequencing strategy:
Zvishandiso: Zvikamu zveDNA zvezvikamu zvina zvemangrove midzi yakabatana samples: ivhu risina kusimwa, rhizosphere, episphere uye endosphere compartments
Platform: Illumina HiSeq 2500
Zvinangwa: Metagenome
16S rRNA gene V3-V4 dunhu
Mibairo yakakosha
Metagenomic sequencing uye metabarcoding profiling pane ivhu-midzi inoenderera yemiti yemangrove yakagadziriswa kuti iongorore kuparadzirwa kwemishonga inorwisa mabhakitiriya (ARGs) kubva muvhu kuenda muzvirimwa.Metagenomic data yakaratidza kuti 91.4% yemishonga inorwisa mabhakitiriya yaiwanzozivikanwa muzvikamu zvina zvevhu zvataurwa pamusoro apa, izvo zvakaratidza fashoni inopfuurira.16S rRNA amplicon sequencing yakagadzira 29,285 sequences, inomiririra 346 marudzi.Kusanganiswa nemhando dzemhando dzemhando neamplicon sequencing, kuparadzirwa uku kwakawanikwa kwakasununguka kubva kune midzi-inosanganiswa microbiota, zvisinei, inogona kufambiswa nenharembozha yemajini.Ongororo iyi yakaratidza kuyerera kweArGs uye utachiona kubva muvhu kupinda muzvirimwa kuburikidza nekubatana kwevhu-midzi inoenderera.
Reference
Wang, C. , Hu, R. , Strong, PJ , Zhuang, W. , & Shu, L. .(2020).Kuwanda kwemajini ekudzivirira kwemishonga inorwisa mabhakitiriya uye hutachiona hwehutachiona padyo nevhu-mangrove midzi inoenderera.Nyaya yeHazardous Materials, 408, 124985.